4USJ
 
 | | N-acetylglutamate kinase from Arabidopsis thaliana in complex with PII from Chlamydomonas reinhardtii | | 分子名称: | ACETYLGLUTAMATE KINASE, CHLOROPLASTIC, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Chellamuthu, V.R, Forchhammer, K, Hartmann, M.D. | | 登録日 | 2014-07-08 | | 公開日 | 2014-12-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A Widespread Glutamine-Sensing Mechanism in the Plant Kingdom.
Cell(Cambridge,Mass.), 159, 2014
|
|
4W74
 
 | |
4USA
 
 | | Aldehyde Oxidoreductase from Desulfovibrio gigas (MOP), soaked with trans-cinnamaldehyde | | 分子名称: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, BICARBONATE ION, ... | | 著者 | Correia, H.D, Romao, M.J, Santos-Silva, T. | | 登録日 | 2014-07-03 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Aromatic Aldehydes at the Active Site of Aldehyde Oxidoreductase from Desulfovibrio Gigas: Reactivity and Molecular Details of the Enzyme-Substrate and Enzyme-Product Interaction.
J.Biol.Inorg.Chem., 20, 2015
|
|
4UWQ
 
 | | Crystal structure of the disulfide-linked complex of the thiosulfodyrolase SoxB with the carrier-protein SoxYZ from Thermus thermophilus | | 分子名称: | MANGANESE (II) ION, SOXY PROTEIN, SOXZ, ... | | 著者 | Grabarczyk, D.B, Chappell, P.E, Johnson, S, Stelzl, L.S, Lea, S.M, Berks, B.C. | | 登録日 | 2014-08-14 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.28 Å) | | 主引用文献 | Structural Basis for Specificity and Promiscuity in a Carrier Protein/Enzyme System from the Sulfur Cycle
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4V9R
 
 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | 著者 | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | 登録日 | 2013-12-05 | | 公開日 | 2014-07-09 | | 最終更新日 | 2014-12-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4W2H
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pactamycin (co-crystallized), mRNA and deacylated tRNA in the P site | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | 著者 | Polikanov, Y.S, Osterman, I.A, Szal, T, Tashlitsky, V.N, Serebryakova, M.V, Kusochek, P, Bulkley, D, Malanicheva, I.A, Efimenko, T.A, Efremenkova, O.V, Konevega, A.L, Shaw, K.J, Bogdanov, A.A, Rodnina, M.V, Dontsova, O.A, Mankin, A.S, Steitz, T.A, Sergiev, P.V. | | 登録日 | 2014-09-12 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome.
Mol.Cell, 56, 2014
|
|
4V4P
 
 | | Crystal structure of 70S ribosome with thrS operator and tRNAs. | | 分子名称: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Jenner, L, Romby, P, Rees, B, Schulze-Briese, C, Springer, M, Ehresmann, C, Ehresmann, B, Moras, D, Yusupova, G, Yusupov, M. | | 登録日 | 2005-01-19 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (5.5 Å) | | 主引用文献 | Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding.
Science, 308, 2005
|
|
4V2U
 
 | | Apo-structure of alpha2,3-sialyltransferase from Pasteurella dagmatis | | 分子名称: | SIALYLTRANSFERASE | | 著者 | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | 登録日 | 2014-10-15 | | 公開日 | 2015-04-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4UW7
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber without its intra-molecular chaperone domain | | 分子名称: | GLYCEROL, L-SHAPED TAIL FIBER PROTEIN | | 著者 | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | 登録日 | 2014-08-08 | | 公開日 | 2015-08-05 | | 最終更新日 | 2018-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
4V54
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with ribosome recycling factor (RRF). | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | 登録日 | 2007-06-16 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V9Q
 
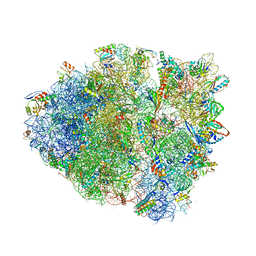 | | Crystal Structure of Blasticidin S Bound to Thermus Thermophilus 70S Ribosome. | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Svidritskiy, E, Ling, C, Ermolenko, D.N, Korostelev, A.A. | | 登録日 | 2013-06-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4UUU
 
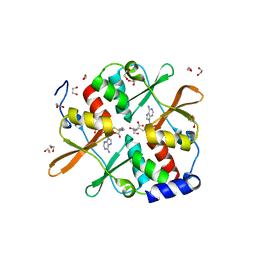 | | 1.7 A resolution structure of human cystathionine beta-synthase regulatory domain (del 516-525) in complex with SAM | | 分子名称: | 1,2-ETHANEDIOL, CYSTATHIONINE BETA-SYNTHASE, S-ADENOSYLMETHIONINE | | 著者 | Kopec, J, McCorvie, T.J, Fitzpatrick, F, Strain-Damerell, C, Froese, D.S, Tallant, C, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | 登録日 | 2014-07-31 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Inter-Domain Communication of Human Cystathionine Beta Synthase: Structural Basis of S-Adenosyl-L-Methionine Activation.
J.Biol.Chem., 289, 2014
|
|
4V2K
 
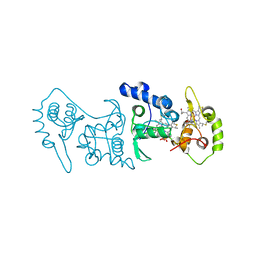 | | Crystal structure of the thiosulfate dehydrogenase TsdA in complex with thiosulfate | | 分子名称: | HEME C, THIOSULFATE, THIOSULFATE DEHYDROGENASE | | 著者 | Grabarczyk, D.B, Chappell, P.E, Eisel, B, Johnson, S, Lea, S.M, Berks, B.C. | | 登録日 | 2014-10-10 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Mechanism of Thiosulfate Oxidation in the Soxa Family of Cysteine-Ligated Cytochromes
J.Biol.Chem., 290, 2015
|
|
4V0I
 
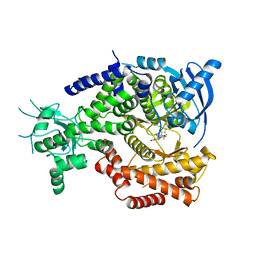 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | 分子名称: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | 著者 | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | 登録日 | 2014-09-16 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
4V24
 
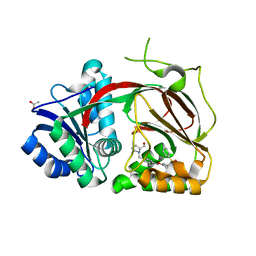 | | Sphingosine kinase 1 in complex with PF-543 | | 分子名称: | ACETATE ION, SPHINGOSINE KINASE 1, {(2R)-1-[4-({3-METHYL-5-[(PHENYLSULFONYL)METHYL]PHENOXY}METHYL)BENZYL]PYRROLIDIN-2-YL}METHANOL | | 著者 | Elkins, J.M, Wang, J, Sorrell, F, Tallant, C, Wang, D, Shrestha, L, Bountra, C, von Delft, F, Knapp, S, Edwards, A. | | 登録日 | 2014-10-05 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Sphingosine Kinase 1 with Pf-543.
Acs Med.Chem.Lett., 5, 2014
|
|
4V61
 
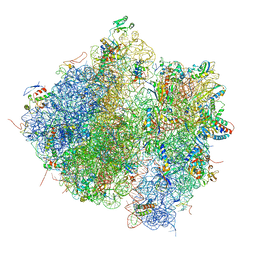 | | Homology model for the Spinach chloroplast 30S subunit fitted to 9.4A cryo-EM map of the 70S chlororibosome. | | 分子名称: | 16S rRNA, 23S rRNA, 4.8S rRNA, ... | | 著者 | Sharma, M.R, Wilson, D.N, Datta, P.P, Barat, C, Schluenzen, F, Fucini, P, Agrawal, R.K. | | 登録日 | 2007-11-09 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (9.4 Å) | | 主引用文献 | Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4V6B
 
 | |
4V9H
 
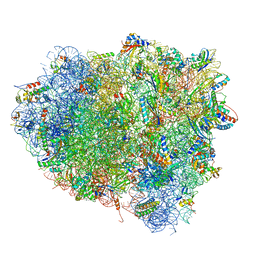 | | Crystal structure of the ribosome bound to elongation factor G in the guanosine triphosphatase state | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Tourigny, D.S, Fernandez, I.S, Kelley, A.C, Ramakrishnan, V. | | 登録日 | 2013-03-25 | | 公開日 | 2014-07-09 | | 最終更新日 | 2014-12-10 | | 実験手法 | X-RAY DIFFRACTION (2.857 Å) | | 主引用文献 | Elongation factor G bound to the ribosome in an intermediate state of translocation.
Science, 340, 2013
|
|
7NXD
 
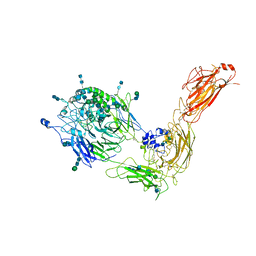 | | Cryo-EM structure of human integrin alpha5beta1 in the half-bent conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Schumacher, S, Dedden, D, Vazquez Nunez, R, Matoba, K, Takagi, J, Biertumpfel, C, Mizuno, N. | | 登録日 | 2021-03-18 | | 公開日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural insights into integrin alpha 5 beta 1 opening by fibronectin ligand.
Sci Adv, 7, 2021
|
|
7NYD
 
 | | cryoEM structure of 2C9-sMAC | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Menny, A, Couves, E.C, Bubeck, D. | | 登録日 | 2021-03-22 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Structural basis of soluble membrane attack complex packaging for clearance.
Nat Commun, 12, 2021
|
|
7NYC
 
 | | cryoEM structure of 3C9-sMAC | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Menny, A, Couves, E.C, Bubeck, D. | | 登録日 | 2021-03-22 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Structural basis of soluble membrane attack complex packaging for clearance.
Nat Commun, 12, 2021
|
|
4V4Q
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli at 3.5 A resolution. | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Schuwirth, B.S, Borovinskaya, M.A, Hau, C.W, Zhang, W, Vila-Sanjurjo, A, Holton, J.M, Cate, J.H.D. | | 登録日 | 2005-08-30 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.46 Å) | | 主引用文献 | Structures of the bacterial ribosome at 3.5 A resolution.
Science, 310, 2005
|
|
7NXV
 
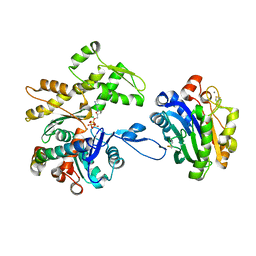 | | Crystal structure of the complex of DNase I/G-actin/PPP1R15A_582-621 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | 著者 | Yan, Y, Ron, D. | | 登録日 | 2021-03-19 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Higher-order phosphatase-substrate contacts terminate the integrated stress response.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OJP
 
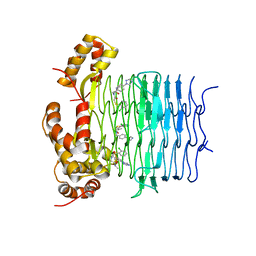 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 1 | | 分子名称: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, MAGNESIUM ION | | 著者 | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | 登録日 | 2021-05-17 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OK2
 
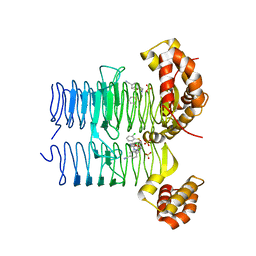 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 3 | | 分子名称: | Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, SULFATE ION, ~{N}-[(5-azanyl-1,3,4-oxadiazol-2-yl)methyl]-2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]ethanamide | | 著者 | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | 登録日 | 2021-05-17 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
