6R1D
 
 | |
5LCX
 
 | | Isopiperitenone reductase from Mentha piperita in complex with NADP | | Descriptor: | (-)-isopiperitenone reductase, (4S)-2-METHYL-2,4-PENTANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7SEZ
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.70001245 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
7SF0
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with trinucleotide substrate | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, MAGNESIUM ION, ... | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95000446 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
6R1K
 
 | |
8BQ3
 
 | |
5F1U
 
 | | biomimetic design results in a potent allosteric inhibitor of dihydrodipicolinate synthase from Campylobacter jejuni | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Conly, C.J.T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Biomimetic Design Results in a Potent Allosteric Inhibitor of Dihydrodipicolinate Synthase from Campylobacter jejuni.
J.Am.Chem.Soc., 138, 2016
|
|
6R1W
 
 | |
6R1X
 
 | |
5LSG
 
 | | PPARgamma complex with the betulinic acid | | Descriptor: | Betulinic Acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Calleri, E, Paiardini, A. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Betulinic acid is a PPAR gamma antagonist that improves glucose uptake, promotes osteogenesis and inhibits adipogenesis.
Sci Rep, 7, 2017
|
|
7SKI
 
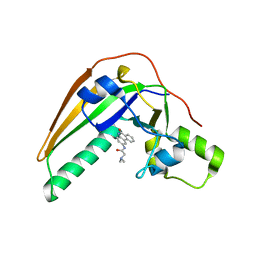 | | Pertussis toxin in complex with PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Pertussis toxin subunit 1 | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2021-10-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.09912443 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
8G95
 
 | |
6R1C
 
 | |
6U4V
 
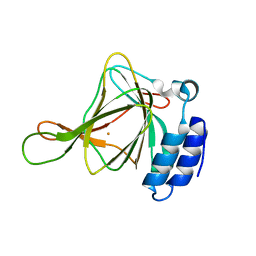 | | Non-crosslinked wild type cysteine dioxygenase | | Descriptor: | Cysteine dioxygenase type 1, FE (III) ION | | Authors: | Meneely, K.M, Chilton, A.S, Forbes, D.L, Ellis, H.R, Lamb, A.L. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 3-His Metal Coordination Site Promotes the Coupling of Oxygen Activation to Cysteine Oxidation in Cysteine Dioxygenase.
Biochemistry, 59, 2020
|
|
5LW6
 
 | |
7RZW
 
 | | CryoEM structure of Arabidopsis thaliana phytochrome B | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B | | Authors: | Li, H, Burgie, E.S, Vierstra, R.D, Li, H. | | Deposit date: | 2021-08-27 | | Release date: | 2022-04-13 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Plant phytochrome B is an asymmetric dimer with unique signalling potential.
Nature, 604, 2022
|
|
6R3O
 
 | | Aspergillus niger ferric acid decarboxylase (Fdc) L439G variant in complex with prFMN (purified in the radical form) and phenylpropiolic acid | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D. | | Deposit date: | 2019-03-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Atomic description of an enzyme reaction dependent on reversible 1,3-dipolar cycloaddition
to be published
|
|
6R42
 
 | |
6U7O
 
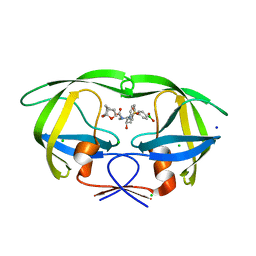 | | HIV-1 wild type protease with GRL-00819A, with phenyl-boronic-acid as P2'-ligand and with a 6-5-5-ring fused crown-like tetrahydropyranofuran as the P2-ligand | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Kneller, D.W, Weber, I.T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Potent HIV-1 Protease Inhibitors Containing Carboxylic and Boronic Acids: Effect on Enzyme Inhibition and Antiviral Activity and Protein-Ligand X-ray Structural Studies.
Chemmedchem, 14, 2019
|
|
7SD0
 
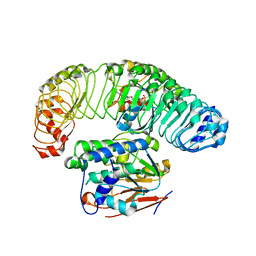 | | Cryo-EM structure of the SHOC2:PP1C:MRAS complex | | Descriptor: | Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Liau, N.P.D, Johnson, M.C, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for SHOC2 modulation of RAS signalling.
Nature, 609, 2022
|
|
5XF2
 
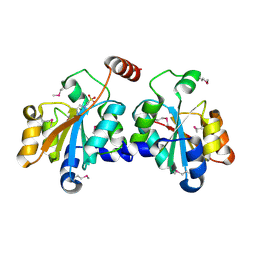 | | Crystal structure of SeMet-HldC from Burkholderia pseudomallei | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-04-07 | | Release date: | 2017-07-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Expression and crystallographic studies of D-glycero-beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5X61
 
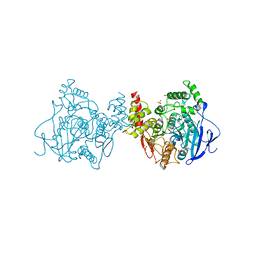 | | Crystal structure of Acetylcholinesterase Catalytic Subunit of the Malaria Vector Anopheles Gambiae, 3.4 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, ... | | Authors: | Han, Q, Robinson, H, Ding, H, Wong, D.M, Lam, P.C.H, Totrov, M.M, Carlier, P.R, Li, J. | | Deposit date: | 2017-02-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector Anopheles gambiae
Insect Sci., 2017
|
|
6U07
 
 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
3GTM
 
 | | Co-complex of Backtracked RNA polymerase II with TFIIS | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
7SCW
 
 | | KRAS full length wild-type in complex with RGL1 Ras association domain | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Eves, B.J, Kuntz, D.A, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-09-29 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of RGL1 RAS-Association Domain in Complex with KRAS and the Oncogenic G12V Mutant.
J.Mol.Biol., 434, 2022
|
|
