5DU1
 
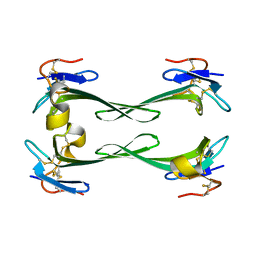 | | Crystal structure of Dendroaspis polylepis mambalgin-1 wild-type in P21 space group. | | Descriptor: | Mambalgin-1 | | Authors: | Stura, E.A, Tepshi, L, Mourier, G, Kessler, P, Servent, D. | | Deposit date: | 2015-09-18 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
4FAM
 
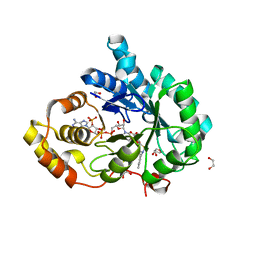 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with 3-((3,4-dihydroisoquinolin-2(1H)-yl)sulfonyl)benzoic acid (17) | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,4-dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acid, Aldo-keto reductase family 1 member C3, ... | | Authors: | Turnbull, A.P, Jamieson, S.M.F, Brooke, D.G, Heinrich, D, Atwell, G.J, Silva, S, Hamilton, E.J, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Flanagan, J.U, Denny, W.A. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids; a New Class of Highly Potent and Selective Inhibitors of the Type 5 17-beta-hydroxysteroid Dehydrogenase AKR1C3
J.Med.Chem., 55, 2012
|
|
2XBQ
 
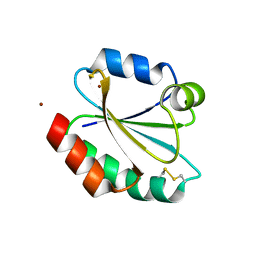 | | Crystal structure of reduced Schistosoma mansoni Thioredoxin pre- protein at 1.7 Angstrom | | Descriptor: | THIOREDOXIN, ZINC ION | | Authors: | Boumis, G, Miele, A.E, Dimastrogiovanni, D, Angelucci, F, Bellelli, A. | | Deposit date: | 2010-04-14 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Functional Characterization of Schistosoma Mansoni Thioredoxin.
Protein Sci., 20, 2011
|
|
4V41
 
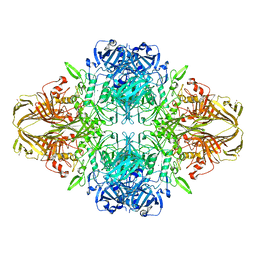 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
2G8T
 
 | |
2XWQ
 
 | | Anaerobic cobalt chelatase from Archeaoglobus fulgidus (CbiX) in complex with metalated sirohydrochlorin product | | Descriptor: | COBALT SIROHYDROCHLORIN, SIROHYDROCHLORIN COBALTOCHELATASE | | Authors: | Ladakis, D, Brindley, A.A, Deery, E, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2010-11-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5EA5
 
 | | Crystal Structure of Inhibitor TMC-353121 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-[[6-[[[2-(3-hydroxypropyl)-5-methylphenyl]amino]methyl]-2-[[3-(4-morpholinyl)propyl]amino]-1H-benzimidazol-1-yl]methyl]-6-methyl-3-pyridinol, D(-)-TARTARIC ACID, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
2XBR
 
 | | Raman crystallography of Hen White Egg Lysozyme - Low X-ray dose (0.2 MGy) | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Carpentier, P, Royant, A, Weik, M, Bourgeois, D. | | Deposit date: | 2010-04-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Raman Assisted Crystallography Reveals a Mechanism of X-Ray Induced Reversible Disulfide Radical Formation
Structure, 18, 2010
|
|
4OUN
 
 | | Crystal Structure of Mini-ribonuclease 3 from Bacillus subtilis | | Descriptor: | Mini-ribonuclease 3 | | Authors: | Chojnowski, G, Czarnecka, J, Nowak, E, Pianka, D, Glow, D, Sabala, I, Skowronek, K, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2014-02-18 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence-specific cleavage of dsRNA by Mini-III RNase
Nucleic Acids Res., 43, 2015
|
|
6R40
 
 | |
4F64
 
 | | Crystal structure of Human Fibroblast Growth Factor Receptor 1 Kinase domain in complex with compound 6 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N~4~-[3-(3-methoxypropyl)-1H-pyrazol-5-yl]-N~2~-[(3-methyl-1,2-oxazol-5-yl)methyl]pyrimidine-2,4-diamine, Fibroblast growth factor receptor 1, ... | | Authors: | Norman, R.A, Breed, J, Ogg, D. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein-Ligand Crystal Structures Can Guide the Design of Selective Inhibitors of the FGFR Tyrosine Kinase.
J.Med.Chem., 55, 2012
|
|
4F63
 
 | | Crystal structure of Human Fibroblast Growth Factor Receptor 1 Kinase domain in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N~4~-(3-methyl-1H-pyrazol-5-yl)-N~2~-[2-(pyridin-3-yl)ethyl]pyrimidine-2,4-diamine, Fibroblast growth factor receptor 1 | | Authors: | Norman, R.A, Breed, J, Ogg, D. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Protein-Ligand Crystal Structures Can Guide the Design of Selective Inhibitors of the FGFR Tyrosine Kinase.
J.Med.Chem., 55, 2012
|
|
4EZ4
 
 | | free KDM6B structure | | Descriptor: | Lysine-specific demethylase 6B, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Cheng, Z.J, Patel, D.J. | | Deposit date: | 2012-05-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response.
Nature, 488, 2012
|
|
6X1K
 
 | | Solution NMR structure of de novo designed TMB2.3 | | Descriptor: | De novo designed transmembrane beta-barrel TMB2.3 | | Authors: | Liang, B, Vorobieva, A.A, Chow, C.M, Baker, D, Tamm, L.K. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of transmembrane beta barrels.
Science, 371, 2021
|
|
4FAL
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with 3-((3,4-dihydroisoquinolin-2(1H)-yl)sulfonyl)-N-methylbenzamide (80) | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,4-dihydroisoquinolin-2(1H)-ylsulfonyl)-N-methylbenzamide, Aldo-keto reductase family 1 member C3, ... | | Authors: | Turnbull, A.P, Jamieson, S.M.F, Brooke, D.G, Heinrich, D, Atwell, G.J, Silva, S, Hamilton, E.J, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Flanagan, J.U, Denny, W.A. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids; a New Class of Highly Potent and Selective Inhibitors of the Type 5 17-beta-hydroxysteroid Dehydrogenase AKR1C3
J.Med.Chem., 55, 2012
|
|
3KO0
 
 | | Structure of the tfp-ca2+-bound activated form of the s100a4 Metastasis factor | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, Protein S100-A4 | | Authors: | Malashkevich, V.N, Dulyaninova, N.G, Knight, D, Almo, S.C, Bresnick, A.R. | | Deposit date: | 2009-11-12 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenothiazines inhibit S100A4 function by inducing protein oligomerization.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5KMS
 
 | |
4O6O
 
 | | Structural and functional studies the characterization of Cys4 Zinc-finger motif in the recombination mediator protein RecR | | Descriptor: | IMIDAZOLE, Recombination protein RecR, ZINC ION | | Authors: | Tang, Q, Liu, Y.P, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-12-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of Cys4 zinc finger motif in the recombination mediator protein RecR.
DNA Repair (Amst.), 24, 2014
|
|
1I4O
 
 | | CRYSTAL STRUCTURE OF THE XIAP/CASPASE-7 COMPLEX | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 4, CASPASE-7 | | Authors: | Huang, Y, Park, Y.C, Rich, R.L, Segal, D, Myszka, D.G, Wu, H. | | Deposit date: | 2001-02-22 | | Release date: | 2001-03-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of caspase inhibition by XIAP: differential roles of the linker versus the BIR domain.
Cell(Cambridge,Mass.), 104, 2001
|
|
6R42
 
 | |
4F9D
 
 | | Structure of Escherichia coli PgaB 42-655 in complex with nickel | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Little, D.J, Poloczek, J, Whitney, J.C, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2012-05-18 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure and Metal Dependent Activity of Escherichia coli PgaB Provides Insight into the Partial De-N-acetylation of Poly-b-1,6-N-acetyl-D-glucosamine
J.Biol.Chem., 287, 2012
|
|
5KRI
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 16b-benzyl 17b-estradiol | | Descriptor: | (8~{R},9~{S},13~{S},14~{S},16~{R},17~{S})-13-methyl-16-(phenylmethyl)-6,7,8,9,11,12,14,15,16,17-decahydrocyclopenta[a]phenanthrene-3,17-diol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
4F3T
 
 | | Human Argonaute-2 - miR-20a complex | | Descriptor: | PHENOL, Protein argonaute-2, RNA (5'-R(P*UP*AP*AP*AP*GP*UP*GP*CP*UP*UP*AP*UP*AP*GP*UP*G*CP*AP*GP*G)-3') | | Authors: | Elkayam, E, Kuhn, C.-D, Tocilj, A, Joshua-Tor, L. | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of Human Argonaute-2 in Complex with miR-20a.
Cell(Cambridge,Mass.), 150, 2012
|
|
6XAG
 
 | | Apo BRAF dimer bound to 14-3-3 | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Liau, N.P.D, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2020-06-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dimerization Induced by C-Terminal 14-3-3 Binding Is Sufficient for BRAF Kinase Activation.
Biochemistry, 59, 2020
|
|
5DO6
 
 | | Crystal structure of Dendroaspis polylepis venom mambalgin-1 T23A mutant | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Mambalgin-1, ... | | Authors: | Stura, E.A, Tepshi, L, Kessler, P, Gilles, M, Servent, D. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-30 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
