1LDN
 
 | | STRUCTURE OF A TERNARY COMPLEX OF AN ALLOSTERIC LACTATE DEHYDROGENASE FROM BACILLUS STEAROTHERMOPHILUS AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, L-LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Wigley, D.B, Gamblin, S.J, Turkenburg, J.P, Dodson, E.J, Piontek, K, Muirhead, H, Holbrook, J.J. | | Deposit date: | 1991-11-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a ternary complex of an allosteric lactate dehydrogenase from Bacillus stearothermophilus at 2.5 A resolution.
J.Mol.Biol., 223, 1992
|
|
1B04
 
 | |
4V41
 
 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
5OAF
 
 | | Human Rvb1/Rvb2 heterohexamer in INO80 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Aramayo, R.J, Bythell-Douglas, R, Ayala, R, Willhoft, O, Wigley, D, Zhang, X. | | Deposit date: | 2017-06-21 | | Release date: | 2017-12-13 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Cryo-EM structures of the human INO80 chromatin-remodeling complex.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1A0I
 
 | | ATP-DEPENDENT DNA LIGASE FROM BACTERIOPHAGE T7 COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA LIGASE | | Authors: | Subramanya, H.S, Doherty, A.J, Ashford, S.R, Wigley, D.B. | | Deposit date: | 1997-12-01 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7.
Cell(Cambridge,Mass.), 85, 1996
|
|
1A0P
 
 | | SITE-SPECIFIC RECOMBINASE, XERD | | Descriptor: | SITE-SPECIFIC RECOMBINASE XERD | | Authors: | Subramanya, H.S, Arciszewska, L.K, Baker, R.A, Bird, L.E, Sherratt, D.J, Wigley, D.B. | | Deposit date: | 1997-12-05 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the site-specific recombinase, XerD.
EMBO J., 16, 1997
|
|
2IGD
 
 | |
6HTS
 
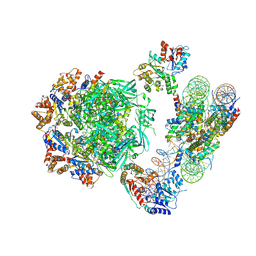 | | Cryo-EM structure of the human INO80 complex bound to nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Ayala, R, Willhoft, O, Aramayo, R.J, Wilkinson, M, McCormack, E.A, Ocloo, L, Wigley, D.B, Zhang, X. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and regulation of the human INO80-nucleosome complex.
Nature, 556, 2018
|
|
3PJR
 
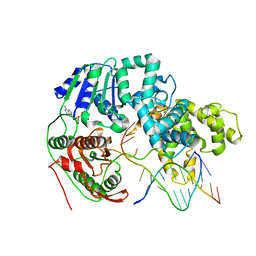 | | HELICASE SUBSTRATE COMPLEX | | Descriptor: | 5'-D(*CP*GP*AP*GP*CP*AP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*TP*GP*CP*TP*CP*GP*TP*TP*TP*TP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Velankar, S.S, Soultanas, P, Dillingham, M.S, Subramanya, H.S, Wigley, D.B. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism
Cell(Cambridge,Mass.), 97, 1999
|
|
3E1S
 
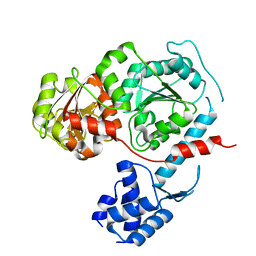 | | Structure of an N-terminal truncation of Deinococcus radiodurans RecD2 | | Descriptor: | Exodeoxyribonuclease V, subunit RecD | | Authors: | Saikrishnan, K, Griffiths, S.P, Cook, N, Court, R, Wigley, D.B. | | Deposit date: | 2008-08-04 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA binding to RecD: role of the 1B domain in SF1B helicase activity.
Embo J., 27, 2008
|
|
5MBV
 
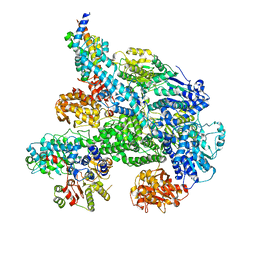 | | Cryo-EM structure of Lambda Phage protein GamS bound to RecBCD. | | Descriptor: | Host-nuclease inhibitor protein gam, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Wilkinson, M, Chaban, Y, Wigley, D.B. | | Deposit date: | 2016-11-08 | | Release date: | 2017-01-11 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the inhibition of RecBCD by Gam and its synergistic antibacterial effect with quinolones.
Elife, 5, 2016
|
|
3U44
 
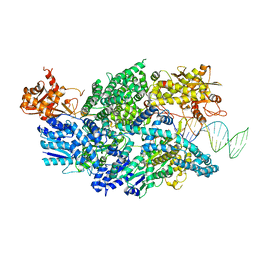 | | Crystal structure of AddAB-DNA complex | | Descriptor: | ATP-dependent helicase/deoxyribonuclease subunit B, ATP-dependent helicase/nuclease subunit A, DNA (36-MER), ... | | Authors: | Saikrishnan, K, Krajewski, W, Wigley, D. | | Deposit date: | 2011-10-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex.
Embo J., 31, 2012
|
|
3U4Q
 
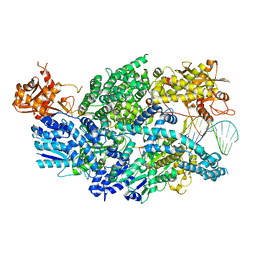 | | Structure of AddAB-DNA complex at 2.8 angstroms | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent helicase/deoxyribonuclease subunit B, ATP-dependent helicase/nuclease subunit A, ... | | Authors: | Saikrishnan, K, Krajewski, W, Wigley, D. | | Deposit date: | 2011-10-10 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex.
Embo J., 31, 2012
|
|
7ZI4
 
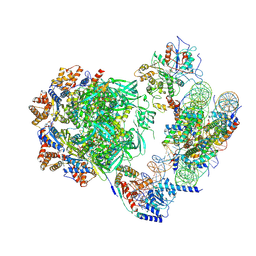 | | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Vance, N.R, Ayala, R, Willhoft, O, Tvardovskiy, A, McCormack, E.A, Bartke, T, Zhang, X, Wigley, D.B. | | Deposit date: | 2022-04-07 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
To Be Published
|
|
3GPL
 
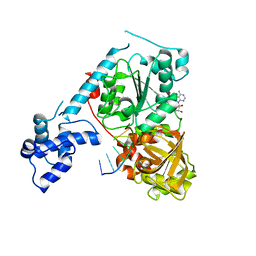 | | Crystal structure of the ternary complex of RecD2 with DNA and ADPNP | | Descriptor: | 5'-D(*T*TP*TP*TP*TP*TP*TP*T)-3', Exodeoxyribonuclease V, subunit RecD, ... | | Authors: | Saikrishnan, K, Cook, N, Wigley, D.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic basis of 5'-3' translocation in SF1B helicases.
Cell(Cambridge,Mass.), 137, 2009
|
|
3K70
 
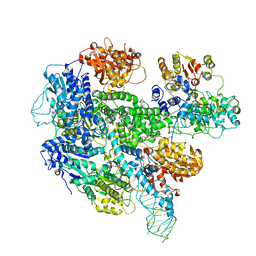 | |
5LD2
 
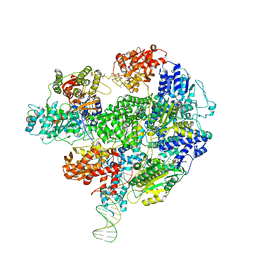 | | Cryo-EM structure of RecBCD+DNA complex revealing activated nuclease domain | | Descriptor: | Fork-Hairpin DNA (70-MER), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wilkinson, M, Chaban, Y, Wigley, D.B. | | Deposit date: | 2016-06-23 | | Release date: | 2016-10-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Mechanism for nuclease regulation in RecBCD.
Elife, 5, 2016
|
|
3GP8
 
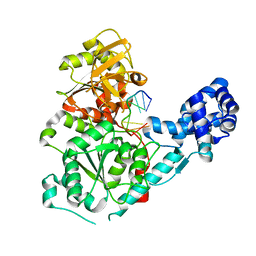 | | Crystal structure of the binary complex of RecD2 with DNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*T*TP*TP*TP*TP*TP*TP*TP*T)-3', Exodeoxyribonuclease V, subunit RecD, ... | | Authors: | Saikrishnan, K, Cook, N, Wigley, D.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic basis of 5'-3' translocation in SF1B helicases.
Cell(Cambridge,Mass.), 137, 2009
|
|
2V1U
 
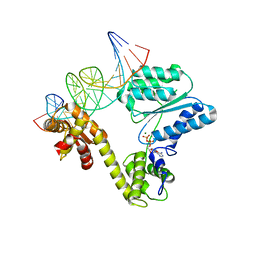 | | STRUCTURE OF THE AEROPYRUM PERNIX ORC1 PROTEIN IN COMPLEX WITH DNA | | Descriptor: | 5'-D(*AP*CP*CP*CP*CP*TP*CP*CP*GP*TP *TP*TP*CP*CP*TP*GP*TP*GP*GP*AP*GP*A)-3', 5'-D(*TP*CP*TP*CP*CP*AP*CP*AP*GP*GP *AP*AP*AP*CP*GP*GP*AP*GP*GP*GP*GP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gaudier, M, Schuwirth, B.S, Westcott, S.L, Wigley, D.B. | | Deposit date: | 2007-05-30 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of DNA Replication Origin Recognition by an Orc Protein.
Science, 317, 2007
|
|
5DMA
 
 | |
6FWS
 
 | |
2PJR
 
 | | HELICASE PRODUCT COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*T)-3'), ... | | Authors: | Velankar, S.S, Soultanas, P, Dillingham, M.S, Subramanya, H.S, Wigley, D.B. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism.
Cell(Cambridge,Mass.), 97, 1999
|
|
2UV1
 
 | |
2UUZ
 
 | |
4CEH
 
 | | Crystal structure of AddAB with a forked DNA substrate | | Descriptor: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | Authors: | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | Deposit date: | 2013-11-11 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
