6XWN
 
 | |
2VMI
 
 | | The structure of seleno-methionine labelled CBM51 from Clostridium perfringens GH95 | | Descriptor: | CALCIUM ION, FIBRONECTIN TYPE III DOMAIN PROTEIN | | Authors: | Gregg, K, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-01-25 | | Release date: | 2008-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
1JF9
 
 | |
6P7O
 
 | | Structure of E. coli MS115-1 NucC, Apo form | | Descriptor: | CHLORIDE ION, E. coli MS115-1 NucC, HEXAETHYLENE GLYCOL, ... | | Authors: | Ye, Q, Lau, R.K, Berg, K.R, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
2VAH
 
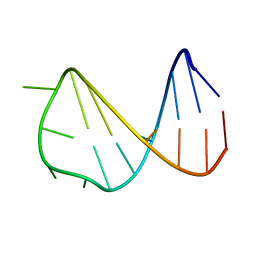 | | Solution structure of a B-DNA hairpin at low pressure. | | Descriptor: | 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*UP*TP *TP*GP*GP*AP*TP*CP*CP*T)-3' | | Authors: | Williamson, M.P, Wilton, D.J, Ghosh, M, Chary, K.V.A, Akasaka, K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Change in a B-DNA Helix with Hydrostatic Pressure
Nucleic Acids Res., 36, 2008
|
|
6MS8
 
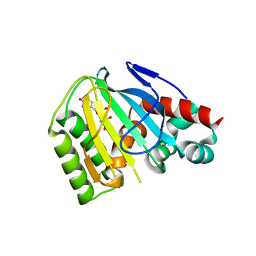 | | Crystal Structure of Chalcone Isomerase from Medicago Truncatula Complexed with (2S) Naringenin | | Descriptor: | Chalcone-flavonone isomerase family protein, NARINGENIN | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-10-16 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
6HBS
 
 | |
2V4X
 
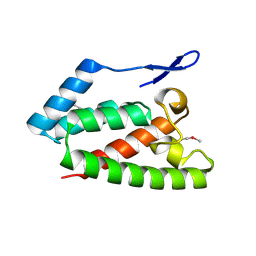 | | Crystal Structure of Jaagsiekte Sheep Retrovirus Capsid N-terminal domain | | Descriptor: | CAPSID PROTEIN P27 | | Authors: | Mortuza, G.B, Goldstone, D.C, Pashley, C, Haire, L.F, Palmarini, M, Taylor, W.R, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Capsid Amino-Terminal Domain from the Betaretrovirus, Jaagsiekte Sheep Retrovirus.
J.Mol.Biol., 386, 2009
|
|
4QU8
 
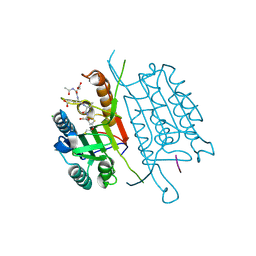 | | Caspase-3 M61A V266H | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CHLORIDE ION, ... | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
5JQA
 
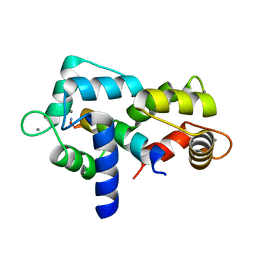 | | CaM:RM20 complex | | Descriptor: | CALCIUM ION, Calmodulin, Myosin light chain kinase, ... | | Authors: | Tokars, V.L, Minasov, G, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2016-05-04 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CaM:RM20 complex
To be Published
|
|
2OD1
 
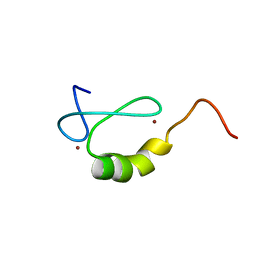 | | Solution structure of the MYND domain from human AML1-ETO | | Descriptor: | Protein CBFA2T1, ZINC ION | | Authors: | Liu, Y.Z, Chen, W, Gaudet, J, Cheney, M.D, Roudaia, L, Cierpicki, T, Klet, R.C, Hartman, K, Laue, T.M, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2006-12-21 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity.
Cancer Cell, 11, 2007
|
|
2V3H
 
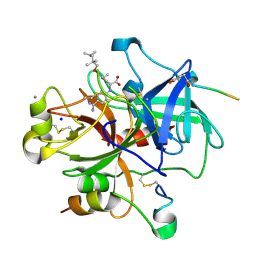 | | Thrombin with 3-cycle no F | | Descriptor: | (2R)-({4-[AMINO(IMINO)METHYL]PHENYL}AMINO){3-[3-(DIMETHYLAMINO)-2,2-DIMETHYLPROPOXY]-5-ETHYLPHENYL}ACETIC ACID, CALCIUM ION, HIRUDIN IIA, ... | | Authors: | Banner, D.W, Obst, U. | | Deposit date: | 2007-06-18 | | Release date: | 2008-06-24 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fluorine in Pharmaceuticals: Looking Beyond Intuition.
Science, 317, 2007
|
|
1XOA
 
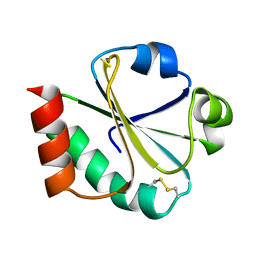 | | THIOREDOXIN (OXIDIZED DISULFIDE FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
6HBV
 
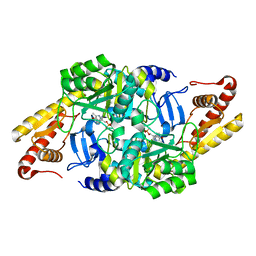 | |
6N3Z
 
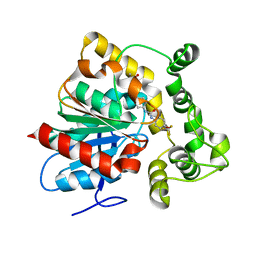 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 4 | | Descriptor: | Epoxide hydrolase TrEH, N-methyl-4-{[trans-4-({[4-(trifluoromethoxy)phenyl]carbamoyl}amino)cyclohexyl]oxy}benzamide | | Authors: | de Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
2W0G
 
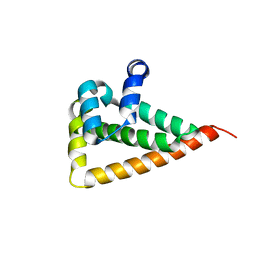 | |
6HQV
 
 | | Pentafunctional AROM Complex from Chaetomium thermophilum | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, GLUTAMIC ACID, ... | | Authors: | Arora Verasto, H, Hartmann, M.D. | | Deposit date: | 2018-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Architecture and functional dynamics of the pentafunctional AROM complex.
Nat.Chem.Biol., 16, 2020
|
|
6V4A
 
 | |
6MSH
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
1XQ6
 
 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g02240 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, unknown protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-11 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g02240
To be published
|
|
6UZT
 
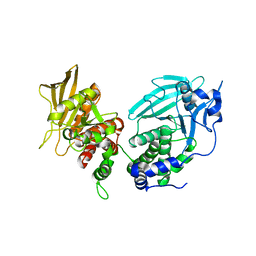 | | Crystal Structure of RPTP alpha | | Descriptor: | Receptor-type tyrosine-protein phosphatase alpha | | Authors: | Santelli, E, Wen, Y, Yang, S, Svensson, M.N.D, Stanford, S.M, Bottini, N. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPTP alpha phosphatase activity is allosterically regulated by the membrane-distal catalytic domain.
J.Biol.Chem., 295, 2020
|
|
6WNX
 
 | | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide | | Descriptor: | Catenin beta-1, F-box/WD repeat-containing protein 11, GLYCEROL, ... | | Authors: | Ivanochko, D, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Boettcher, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-23 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide
To Be Published
|
|
6MVK
 
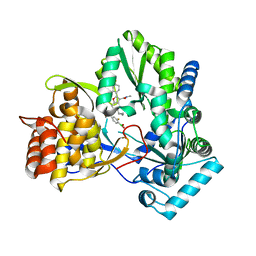 | | HCV NS5B 1b N316 bound to Compound 18 | | Descriptor: | (4-{(4S)-3-[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl]-2-oxo-1,3-oxazolidin-4-yl}-2-fluorophenyl)boronic acid, HCV Polymerase | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
2VSA
 
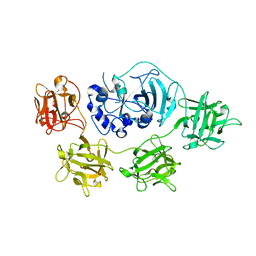 | | Structure and mode of action of a mosquitocidal holotoxin | | Descriptor: | MOSQUITOCIDAL TOXIN | | Authors: | Treiber, N, Reinert, D.J, Carpusca, I, Aktories, K, Schulz, G.E. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure and Mode of Action of a Mosquitocidal Holotoxin
J.Mol.Biol., 381, 2008
|
|
8FC9
 
 | | Cryo-EM structure of the human TRPV4 - RhoA, apo condition | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA, ... | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
