7M7E
 
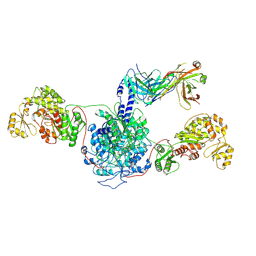 | | 6-Deoxyerythronolide B synthase (DEBS) hybrid module (M3/1) in complex with antibody fragment 1B2 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), 6-deoxyerythronolide-B synthase EryA2, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
7M7G
 
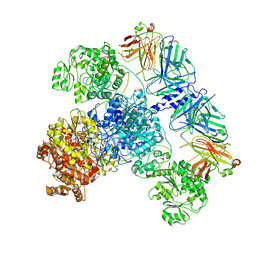 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 2 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
8BQ2
 
 | | CryoEM structure of the pre-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Appleby, R, Bollschweiler, D, Pellegrini, L. | | Deposit date: | 2022-11-18 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly.
Iscience, 26, 2023
|
|
7M7H
 
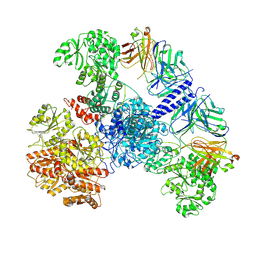 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 1' | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
6OIZ
 
 | | Amyloid-Beta (20-34) wild type | | Descriptor: | Amyloid beta A4 protein | | Authors: | Sawaya, M.R, Warmack, R.A, Zee, C.T, Gonen, T, Clarke, S.G, Eisenberg, D.S. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Structure of amyloid-beta (20-34) with Alzheimer's-associated isomerization at Asp23 reveals a distinct protofilament interface.
Nat Commun, 10, 2019
|
|
7M7I
 
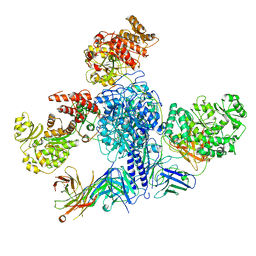 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2 (TE-free) | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
7M7J
 
 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: "turnstile closed" state (TE-free) | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
7RKE
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S in Complex with 4-(((2-chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)amino)methyl)phenol and GRIP Peptide | | Descriptor: | 4-{[(2-chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)amino]methyl}phenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Joiner, C, Sammeta, V.K.R, Norris, J.D, McDonnell, D.P, Willson, T.M, Fanning, S.W. | | Deposit date: | 2021-07-22 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A New Chemotype of Chemically Tractable Nonsteroidal Estrogens Based on a Thieno[2,3- d ]pyrimidine Core.
Acs Med.Chem.Lett., 13, 2022
|
|
8J9V
 
 | | Cryo-EM structure of the African swine fever virus topoisomerase 2 complexed with Cut02aDNA and etoposide (EDI-1) | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*GP*AP*GP*GP*TP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*CP*GP*CP*CP*TP*AP*CP*AP*TP*AP*CP*CP*TP*C)-3'), ... | | Authors: | Chang, C.-W, Tsai, M.-D. | | Deposit date: | 2023-05-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | A unified view on enzyme catalysis by cryo-EM study of a DNA topoisomerase.
Commun Chem, 7, 2024
|
|
8QZJ
 
 | |
7M7F
 
 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 1 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
8J9W
 
 | | Cryo-EM structure of the African swine fever virus topoisomerase 2 complexed with Cut02bDNA and etoposide (EDI-2) | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*AP*AP*GP*AP*AP*CP*TP*CP*TP*GP*TP*AP*G)-3'), DNA (5'-D(*CP*AP*TP*GP*CP*TP*AP*CP*AP*GP*AP*GP*TP*TP*CP*TP*T)-3'), ... | | Authors: | Chang, C.-W, Tsai, M.-D. | | Deposit date: | 2023-05-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A unified view on enzyme catalysis by cryo-EM study of a DNA topoisomerase.
Commun Chem, 7, 2024
|
|
5L7U
 
 | | Crystal structure of BvGH123 with bound GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CHLORIDE ION, Glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
6WFY
 
 | |
8BQF
 
 | | Adenylate Kinase L107I MUTANT | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Scheerer, D, Adkar, B.V, Bhattacharyya, S, Levy, D, Iljina, M, Iljina, I, Dym, O, Haran, G, Shakhnovich, E.I. | | Deposit date: | 2022-11-21 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Allosteric communication between ligand binding domains modulates substrate inhibition in adenylate kinase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6U0O
 
 | | Crystal structure of a peptidoglycan release complex, SagB-SpdC, in lipidic cubic phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-ETHOXYETHOXY)ETHANOL, CITRATE ANION, ... | | Authors: | Owens, T.W, Schaefer, K, Kahne, D, Walker, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization.
Nat Microbiol, 6, 2021
|
|
8BCT
 
 | | X-ray crystal structure of a de novo selected helix-loop-helix heterodimer in a syn arrangement, 26alpha/26beta | | Descriptor: | 26alpha, 26beta, ACETATE ION, ... | | Authors: | Naudin, E.A, Mylemans, B, Smith, A.J, Savery, N.J, Woolfson, D.N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Selection of Heterodimerizing Helical Hairpins for Synthetic Biology.
Acs Synth Biol, 12, 2023
|
|
7R0W
 
 | | 2.8 Angstrom cryo-EM structure of the dimeric cytochrome b6f-PetP complex from Synechocystis sp. PCC 6803 with natively bound lipids and plastoquinone molecules | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S,8E)-1-{[(2S)-1-hydroxy-3-{[(1S)-1-hydroxypentadecyl]oxy}propan-2-yl]oxy}heptadec-8-en-1-ol, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, ... | | Authors: | Farmer, D.F, Proctor, M.S, Malone, L.A, Swainsbury, D.P.K, Hawkings, F.R, Hitchcock, A, Johnson, M.P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of the Synechocystis sp. PCC 6803 cytochrome b6f complex with and without the regulatory PetP subunit.
Biochem.J., 479, 2022
|
|
6WRR
 
 | |
6OF8
 
 | | Structure of Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Ile464His, and Phe467Met mutant human CamKII-alpha hub domain | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha, GLYCEROL, POTASSIUM ION | | Authors: | McSpadden, E.D, Chi, C.C, Gee, C.L, Kuriyan, J. | | Deposit date: | 2019-03-28 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variation in assembly stoichiometry in non-metazoan homologs of the hub domain of Ca2+/calmodulin-dependent protein kinase II.
Protein Sci., 28, 2019
|
|
6ZXM
 
 | | Diguanylate cyclase DgcR in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Putative GGDEF/response regulator receiver domain protein | | Authors: | Teixeira, R.D, Schirmer, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Activation mechanism of a small prototypic Rec-GGDEF diguanylate cyclase.
Nat Commun, 12, 2021
|
|
4TGF
 
 | | SOLUTION STRUCTURES OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA DERIVED FROM 1*H NMR DATA | | Descriptor: | DES-VAL-1,VAL-2,TRANSFORMING GROWTH FACTOR, ALPHA | | Authors: | Kline, T.P, Brown, F.K, Brown, S.C, Jeffs, P.W, Kopple, K.D, Mueller, L. | | Deposit date: | 1990-06-13 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human transforming growth factor alpha derived from 1H NMR data.
Biochemistry, 29, 1990
|
|
7ABO
 
 | |
6WP4
 
 | | Pyruvate Kinase M2 mutant-S37E | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nandi, S, Razzaghi, M, Srivastava, D, Dey, M. | | Deposit date: | 2020-04-26 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for allosteric regulation of pyruvate kinase M2 by phosphorylation and acetylation.
J.Biol.Chem., 295, 2020
|
|
6OOT
 
 | | HIV-1 Protease NL4-3 L89V, L90M Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, NL4-3 PROTEASE, SULFATE ION | | Authors: | Henes, M, Kosovrasti, K, Lockbaum, G.J, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A, Whitfield, T.W. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Molecular Determinants of Epistasis in HIV-1 Protease: Elucidating the Interdependence of L89V and L90M Mutations in Resistance.
Biochemistry, 58, 2019
|
|
