6Q38
 
 | | The Crystal structure of CK2a bound to P1-C4 | | Descriptor: | 3,5-bis(1-methyl-1,2,3-triazol-4-yl)benzoic acid, BENZOIC ACID, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, Iegre, J, Baker, D, Tan, Y, Sore, H, Donovan, D, Spring, D, Chandra, V, Hyvonen, M. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Efficient development of stable and highly functionalised peptides targeting the CK2 alpha /CK2 beta protein-protein interaction.
Chem Sci, 10, 2019
|
|
6Q4Q
 
 | | The Crystal structure of CK2a bound to P2-C4 | | Descriptor: | 3,5-bis(1-methyl-1,2,3-triazol-4-yl)benzoic acid, ACETATE ION, BENZOIC ACID, ... | | Authors: | Brear, P, Iegre, J, Baker, D, Tan, Y, Sore, H, Donovan, D, Spring, D, Chandra, V, Hyvonen, M. | | Deposit date: | 2018-12-06 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Efficient development of stable and highly functionalised peptides targeting the CK2 alpha /CK2 beta protein-protein interaction.
Chem Sci, 10, 2019
|
|
8DW2
 
 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Antibody CR3022 heavy chain, Antibody CR3022 light chain, Antibody S309 heavy chain, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
6NTS
 
 | | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | | Descriptor: | MANGANESE (II) ION, N-[(1R,2R,3S)-2-hydroxy-3-(10H-phenoxazin-10-yl)cyclohexyl]-4-(trifluoromethoxy)benzene-1-sulfonamide, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2019-01-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Selective PP2A Enhancement through Biased Heterotrimer Stabilization.
Cell, 181, 2020
|
|
8DW3
 
 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Anti-SARS-CoV-2 DARPin SR16m, Antibody S309 light chain, Spike protein S1, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
4YI3
 
 | | Crystal structure of Gpb in complex with 4a | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Kantsadi, A.L, Chatzileontiadou, D.S.M, Stravodimos, G.A, Leonidas, D.D. | | Deposit date: | 2015-02-27 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycogen phosphorylase as a target for type 2 diabetes: synthetic, biochemical, structural and computational evaluation of novel N-acyl-N -( beta-D-glucopyranosyl) urea inhibitors.
Curr Top Med Chem, 15, 2015
|
|
8QZJ
 
 | |
5LP1
 
 | |
8QZI
 
 | | Crystal structure of PptT-ACP from Mycobacterium tuberculosis | | Descriptor: | 4'-phosphopantetheinyl transferase PptT, COENZYME A, ISOPROPYL ALCOHOL, ... | | Authors: | Gavalda, S, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic Cycle of Type II 4'-Phosphopantetheinyl Transferases
Acs Catalysis, 14, 2024
|
|
7T02
 
 | |
4YDJ
 
 | | Crystal structure of broadly and potently neutralizing antibody 44-VRC13.01 in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
8DXU
 
 | |
8VEI
 
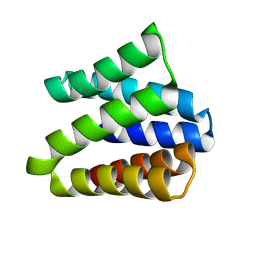 | |
8VEJ
 
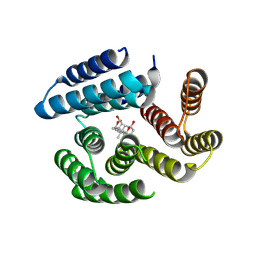 | | De novo designed cholic acid binder: CHD_buttress | | Descriptor: | CHD_buttress, CHOLIC ACID | | Authors: | Bera, A.K, Tran, L, Kang, A, Baker, D. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Binding and sensing diverse small molecules using shape-complementary pseudocycles.
Science, 385, 2024
|
|
8DXT
 
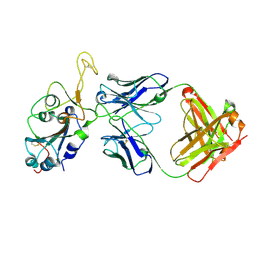 | | Fab arm of antibody GAR12 bound to the receptor binding domain of SARS-CoV-2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab arm of antibody GAR12, Light chain of Fab arm of antibody GAR12, ... | | Authors: | Langley, D.B, Christ, D, Henry, J.Y. | | Deposit date: | 2022-08-03 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Broadly neutralizing SARS-CoV-2 antibodies through epitope-based selection from convalescent patients.
Nat Commun, 14, 2023
|
|
7BBM
 
 | | Mutant nitrobindin M75L/H76L/Q96C/M148L (NB4H) from Arabidopsis thaliana with cofactor MnPPIX | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE PROTOPORPHYRIN IX, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Minges, A, Sauer, D.F, Wittwer, M, Markel, U, Spiertz, M, Schiffels, J, Davari, M.D, Okuda, J, Schwaneberg, U, Groth, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Chemogenetic engineering of nitrobindin toward an artificial epoxygenase
Catalysis Science And Technology, 2021
|
|
8W1V
 
 | | The beta2 adrenergic receptor bound to a bitopic ligand | | Descriptor: | (2S)-1-[(3-{1-[4-(4-{(2S)-2-hydroxy-3-[(propan-2-yl)amino]propoxy}phenyl)butyl]-1H-1,2,3-triazol-4-yl}propyl)amino]-3-(2-propylphenoxy)propan-2-ol, Beta-2 adrenergic receptor,Endolysin, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Gaiser, B, Danielsen, M, Xu, X, Jorgensen, K, Fronik, P, Marcher-Rorsted, E, Wrobe, T, Hirata, K, Liu, X, Mathiesen, J, Pedersen, D. | | Deposit date: | 2024-02-19 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bitopic Ligands Support the Presence of a Metastable Binding Site at the beta 2 Adrenergic Receptor.
J.Med.Chem., 67, 2024
|
|
8QAB
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil hexameric alpha-helical barrel with 4 heptad repeats, apCCHex | | Descriptor: | 1-methoxy-2-[2-[2-[2-[2-[2-[2-[2-[2-[2-(2-methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethane, apCCHex | | Authors: | Naudin, E.N, Albanese, K.I, Petrenas, R, Woolfson, D.N. | | Deposit date: | 2023-08-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rationally seeded computational protein design of ɑ-helical barrels.
Nat.Chem.Biol., 20, 2024
|
|
8Q9Z
 
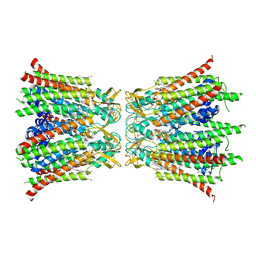 | |
7TJA
 
 | | Structure of the Light Harvesting Complex PE545 from Proteomonas sulcata | | Descriptor: | 15,16-DIHYDROBILIVERDIN, MAGNESIUM ION, PHYCOERYTHROBILIN, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2022-01-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
8QAC
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil 8-helix bundle with 4 heptad repeats, antiparallel 8-helix bundle-GLIA | | Descriptor: | antiparallel 8-helix bundle-GLIA | | Authors: | Albanese, K.I, Dawson, W.M, Petrenas, R, Woolfson, D.N. | | Deposit date: | 2023-08-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rationally seeded computational protein design of ɑ-helical barrels.
Nat.Chem.Biol., 20, 2024
|
|
5N81
 
 | | Crystal structure of an engineered TycA variant in complex with an O-propargyl-beta-Tyr-AMP analog | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Tyrocidine synthase 1, ... | | Authors: | Niquille, D.L, Hansen, D.A, Mori, T, Fercher, D, Kries, H, Hilvert, D. | | Deposit date: | 2017-02-22 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nonribosomal biosynthesis of backbone-modified peptides.
Nat Chem, 10, 2018
|
|
6MXC
 
 | |
7BCA
 
 | |
8EAW
 
 | |
