5AFW
 
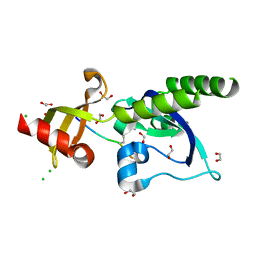 | | Assembly of methylated LSD1 and CHD1 drives AR-dependent transcription and translocation | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 1, ... | | Authors: | Metzger, E, Willmann, D, McMillan, J, Petroll, K, Metzger, P, Gerhardt, S, vonMaessenhausen, A, Schott, A.K, Espejo, A, Eberlin, A, Wohlwend, D, Schuele, K.M, Schleicher, M, Perner, S, Bedford, M.T, Dengjel, J, Flaig, R, Einsle, O, Schuele, R. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Assembly of Methylated Kdm1A and Chd1 Drives Androgen Receptor-Dependent Transcription and Translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4V69
 
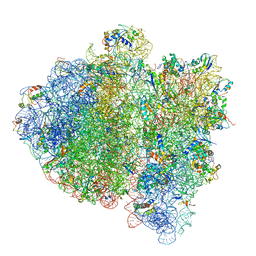 | | Ternary complex-bound E.coli 70S ribosome. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Villa, E, Sengupta, J, Trabuco, L.G, LeBarron, J, Baxter, W.T, Shaikh, T.R, Grassucci, R.A, Nissen, P, Ehrenberg, M, Schulten, K, Frank, J. | | Deposit date: | 2008-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4V6S
 
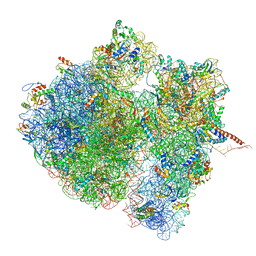 | | Structural characterization of mRNA-tRNA translocation intermediates (class 3 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (13.1 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V6O
 
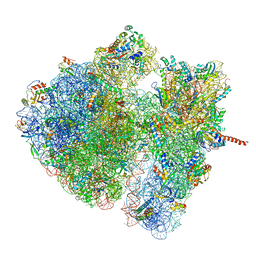 | | Structural characterization of mRNA-tRNA translocation intermediates (class 4a of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7OPB
 
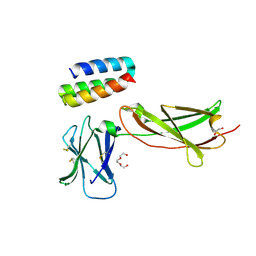 | | IL7R in complex with an antagonist | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IL7R binder, ... | | Authors: | Markovic, I, Verschueren, K.H.G, Verstraete, K, Savvides, S.N. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
6GLW
 
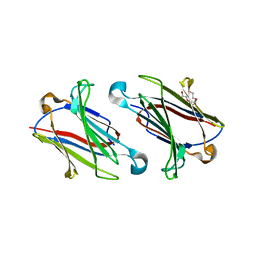 | | Structure of galectin-10 in complex with the Fab fragment of a Charcot-Leyden crystal solubilizing antibody, 1D11 | | Descriptor: | ACETATE ION, Fab 1D11 - Heavy chain, Fab 1D11 - Light chain, ... | | Authors: | Verstraete, K, Verschueren, K, Savvides, S.N. | | Deposit date: | 2018-05-23 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein crystallization promotes type 2 immunity and is reversible by antibody treatment.
Science, 364, 2019
|
|
6GKT
 
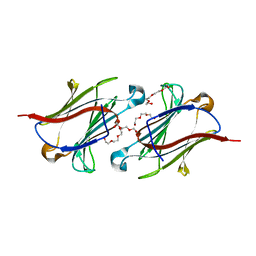 | | X-ray structure of a non-autocrystallizing galectin-10 variant, Gal10-Tyr69Glu | | Descriptor: | Galectin-10, HEXAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | Verstraete, K, Verschueren, K, Savvides, S.N. | | Deposit date: | 2018-05-21 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein crystallization promotes type 2 immunity and is reversible by antibody treatment.
Science, 364, 2019
|
|
6GLX
 
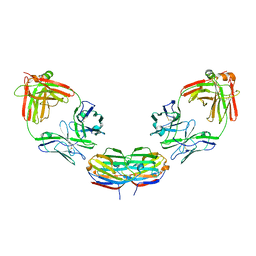 | | Structure of galectin-10 in complex with the Fab fragment of a Charcot-Leyden crystal solubilizing antibody, 4E8 | | Descriptor: | Fab 4E8 - Heavy chain, Fab 4E8 - Light chain, Galectin-10 | | Authors: | Verstraete, K, Verschueren, K, Savvides, S.N. | | Deposit date: | 2018-05-23 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.396 Å) | | Cite: | Protein crystallization promotes type 2 immunity and is reversible by antibody treatment.
Science, 364, 2019
|
|
6GKU
 
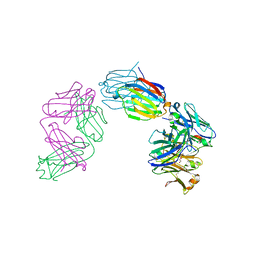 | | Structure of galectin-10 in complex with the Fab fragment of a Charcot-Leyden crystal solubilizing antibody, 6F5 | | Descriptor: | CALCIUM ION, Fab 6F5 - Heavy Chain, Fab 6F5 - Light chain, ... | | Authors: | Verstraete, K, Verschueren, K, Savvides, S.N. | | Deposit date: | 2018-05-21 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Protein crystallization promotes type 2 immunity and is reversible by antibody treatment.
Science, 364, 2019
|
|
6GKQ
 
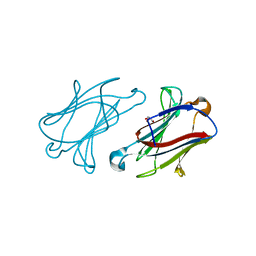 | |
6GKS
 
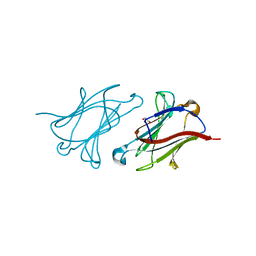 | |
6QCL
 
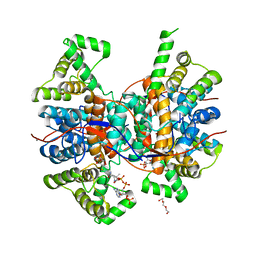 | |
6QFB
 
 | |
5UOT
 
 | |
6HXO
 
 | |
5MPA
 
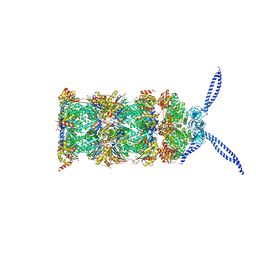 | | 26S proteasome in presence of ATP (s2) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MPC
 
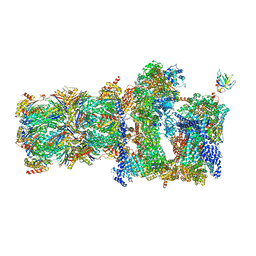 | | 26S proteasome in presence of BeFx (s4) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6HXK
 
 | |
7Z3R
 
 | | Crystal structure of the mouse leptin:LepR-IgCRH2 complex to 2.95 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leptin, Leptin receptor | | Authors: | Verstraete, K, Verschueren, K, Savvides, S.N, Tsirigotaki, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z3P
 
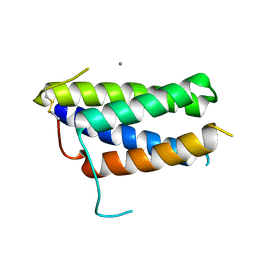 | | Crystal structure of the mouse leptin:LepR-CRH2 encounter complex to 1.95 A resolution. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Leptin, ... | | Authors: | Tsirigotaki, A, Verschueren, K, Savvides, S.N, Verstraete, K. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z3Q
 
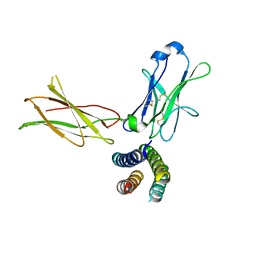 | | Crystal structure of the human leptin:LepR-CRH2 encounter complex to 3.6 A resolution. | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Verschueren, K, Alexandra, T, Savvides, S.N. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.617 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T2T
 
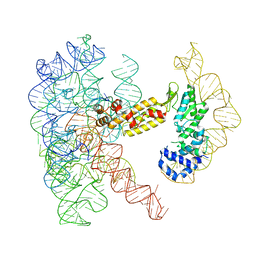 | | Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state | | Descriptor: | AMMONIUM ION, Group II intron reverse transcriptase/maturase, MAGNESIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2R
 
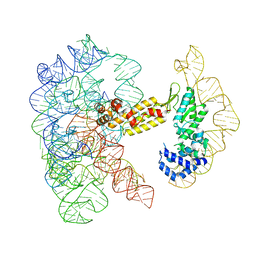 | | Structure of a group II intron ribonucleoprotein in the pre-ligation (pre-2F) state | | Descriptor: | 5'exon, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2S
 
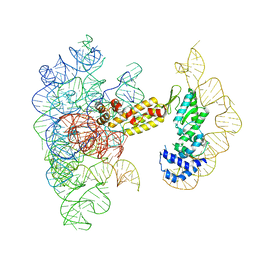 | | Structure of a group II intron ribonucleoprotein in the pre-branching (pre-1F) state | | Descriptor: | AMMONIUM ION, CALCIUM ION, Group II intron reverse transcriptase/maturase, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
6HXL
 
 | |
