7XU1
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU4
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU5
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU6
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU3
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
5T39
 
 | | Crystal Structure of the N-terminal domain of EvdMO1 in the presence of SAH and D-fucose | | Descriptor: | EvdMO1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCulloch, K.M, Starbird, C.A, Chen, Q, Perry, N.A, Berndt, S, Yamakawa, I, Loukachevitch, L.V, Iverson, T.M. | | Deposit date: | 2016-08-25 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1004 Å) | | Cite: | The Structure of the Bifunctional Everninomicin Biosynthetic Enzyme EvdMO1 Suggests Independent Activity of the Fused Methyltransferase-Oxidase Domains.
Biochemistry, 57, 2018
|
|
2MN4
 
 | | NMR solution structure of a computational designed protein based on structure template 1cy5 | | Descriptor: | Computational designed protein based on structure template 1cy5 | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-03-28 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
2MLB
 
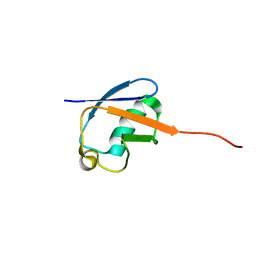 | | NMR solution structure of a computational designed protein based on template of human erythrocytic ubiquitin | | Descriptor: | redesigned ubiquitin | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
5H7O
 
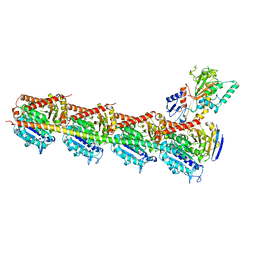 | | Crystal structure of DJ-101 in complex with tubulin protein | | Descriptor: | 2-(1H-indol-4-yl)-4-(3,4,5-trimethoxyphenyl)-1H-imidazo[4,5-c]pyridine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Arnst, K, Wang, Y, Hwang, D.-J, Xue, Y, Costello, T, Hamilton, D, Chen, Q, Yang, J, Park, F, Dalton, J.T, Miller, D.D, Li, W. | | Deposit date: | 2016-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Potent, Metabolically Stable Tubulin Inhibitor Targets the Colchicine Binding Site and Overcomes Taxane Resistance.
Cancer Res., 78, 2018
|
|
5H74
 
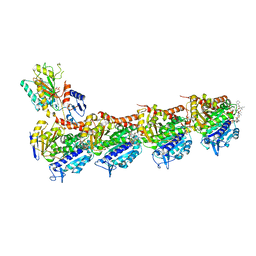 | | Crystal structure of T2R-TTL-14b complex | | Descriptor: | (2~{S},4~{R})-4-[[2-[(1~{R},3~{R})-1-acetyloxy-3-[hexyl-[(2~{S},3~{S})-3-methyl-2-[[(2~{R})-1-methylpiperidin-2-yl]carbonylamino]pentanoyl]amino]-4-methyl-pentyl]-1,3-thiazol-4-yl]carbonylamino]-5-(4-fluorophenyl)-2-methyl-pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2016-11-17 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Activity Relationship Studies of Tubulysin Analogues: Anticancer N-Alkyltubulysins with Subpicomolar Activity and the Crystal Structure Binding to Tubulin
To Be Published
|
|
4O98
 
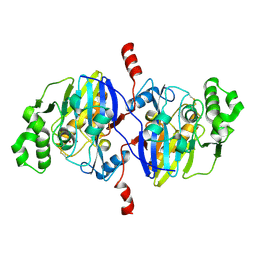 | | Crystal structure of Pseudomonas oleovorans PoOPH mutant H250I/I263W | | Descriptor: | ZINC ION, organophosphorus hydrolase | | Authors: | Luo, X.J, Kong, X.D, Zhao, J, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Switching a newly discovered lactonase into an efficient and thermostable phosphotriesterase by simple double mutations His250Ile/Ile263Trp
Biotechnol.Bioeng., 111, 2014
|
|
4OB6
 
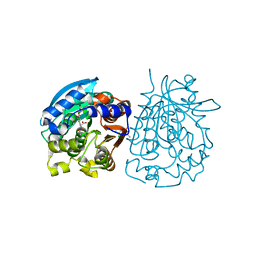 | | Complex structure of esterase rPPE S159A/W187H and substrate (S)-Ac-CPA | | Descriptor: | (2S)-(acetyloxy)(2-chlorophenyl)ethanoic acid, Alpha/beta hydrolase fold-3 domain protein | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4QHD
 
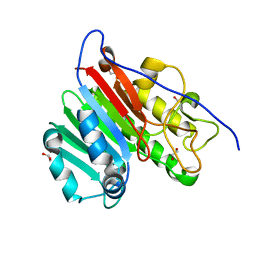 | | Crystal structure of apo human APE1 | | Descriptor: | 1,2-ETHANEDIOL, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | He, H, Chen, Q, Georgiadis, M.M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution crystal structures reveal plasticity in the metal binding site of apurinic/apyrimidinic endonuclease I.
Biochemistry, 53, 2014
|
|
4QHE
 
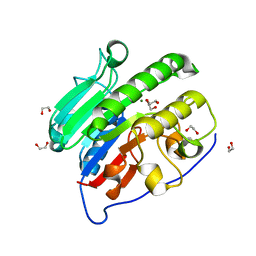 | | Crystal structure of Mg2+ bound human APE1 | | Descriptor: | 1,2-ETHANEDIOL, DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | He, H, Chen, Q, Georgiadis, M.M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structures reveal plasticity in the metal binding site of apurinic/apyrimidinic endonuclease I.
Biochemistry, 53, 2014
|
|
5C8Y
 
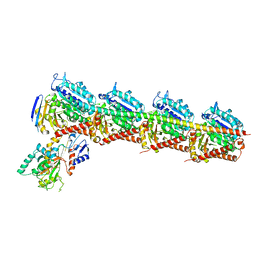 | | Crystal structure of T2R-TTL-Plinabulin complex | | Descriptor: | (3Z,6Z)-3-benzylidene-6-[(5-tert-butyl-1H-imidazol-4-yl)methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-26 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
5CA1
 
 | | Crystal structure of T2R-TTL-Nocodazole complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
5CA0
 
 | | Crystal structure of T2R-TTL-Lexibulin complex | | Descriptor: | 1-ethyl-3-[2-methoxy-4-(5-methyl-4-{[(1S)-1-(pyridin-3-yl)butyl]amino}pyrimidin-2-yl)phenyl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-29 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
5CB4
 
 | | Crystal structure of T2R-TTL-Tivantinib complex | | Descriptor: | (3R,4R)-3-(5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1-yl)-4-(1H-indol-3-yl)pyrrolidine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
5E1J
 
 | | Structure of voltage-gated two-pore channel TPC1 from Arabidopsis thaliana | | Descriptor: | BARIUM ION, CALCIUM ION, Two pore calcium channel protein 1 | | Authors: | Guo, J, Zeng, W, Chen, Q, Lee, C, Chen, L, Yang, Y, Jiang, Y. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.308 Å) | | Cite: | Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana.
Nature, 531, 2016
|
|
5EZY
 
 | | Crystal structure of T2R-TTL-taccalonolide AJ complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-11-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanism of microtubule stabilization by taccalonolide AJ
Nat Commun, 8, 2017
|
|
8HC5
 
 | | SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC8
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with YB13-292 Fab, focused refinement of Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, Light chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HCB
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC7
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) complex with YB9-258 Fab, focused refinement of RBD-dimer region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain variable region of YB9-258, Light chain variable region of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC3
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 2 YB9-258 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
