7E4T
 
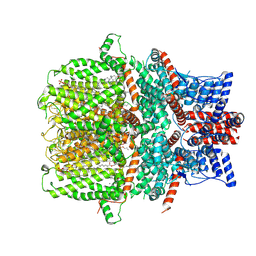 | | Human TRPC5 apo state structure at 3 angstrom | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7D4P
 
 | | Structure of human TRPC5 in complex with clemizole | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-[(4-chlorophenyl)methyl]-2-(pyrrolidin-1-ylmethyl)benzimidazole, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7D4Q
 
 | | Structure of human TRPC5 in complex with HC-070 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 8-(3-chloranylphenoxy)-7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)purine-2,6-dione, CALCIUM ION, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7E28
 
 | |
7CU3
 
 | | Structure of mammalian NALCN-FAM155A complex at 2.65 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, Sodium leak channel non-selective protein, ... | | Authors: | Chen, L, Kang, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-18 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structure of voltage-modulated sodium-selective NALCN-FAM155A channel complex.
Nat Commun, 11, 2020
|
|
7DXF
 
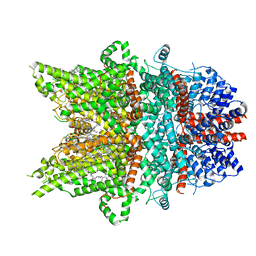 | | Structure of BTDM-bound human TRPC6 nanodisc at 2.9 angstrom in high calcium state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chen, L, Guo, W. | | Deposit date: | 2021-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of human TRPC3 and TRPC6 channel regulation by their intracellular calcium-binding sites.
Neuron, 110, 2022
|
|
7DXC
 
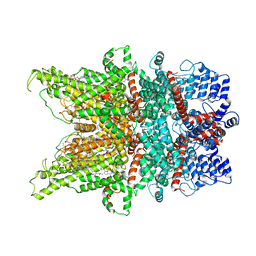 | | Structure of TRPC3 at 3.06 angstrom in low calcium state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL HEMISUCCINATE, Short transient receptor potential channel 3, ... | | Authors: | Chen, L, Guo, W. | | Deposit date: | 2021-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural mechanism of human TRPC3 and TRPC6 channel regulation by their intracellular calcium-binding sites.
Neuron, 110, 2022
|
|
7DXG
 
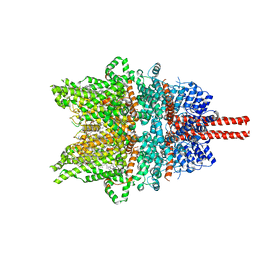 | | Structure of SAR7334-bound TRPC6 at 2.9 angstrom | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[[(1R,2R)-2-[(3R)-3-azanylpiperidin-1-yl]-2,3-dihydro-1H-inden-1-yl]oxy]-3-chloranyl-benzenecarbonitrile, CALCIUM ION, ... | | Authors: | Chen, L, Guo, W. | | Deposit date: | 2021-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of human TRPC3 and TRPC6 channel regulation by their intracellular calcium-binding sites.
Neuron, 110, 2022
|
|
7DXE
 
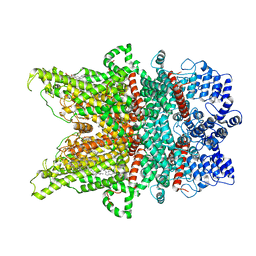 | |
7DXB
 
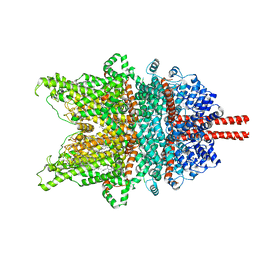 | | Structure of TRPC3 at 2.7 angstrom in high calcium state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chen, L, Guo, W. | | Deposit date: | 2021-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of human TRPC3 and TRPC6 channel regulation by their intracellular calcium-binding sites.
Neuron, 110, 2022
|
|
7D3E
 
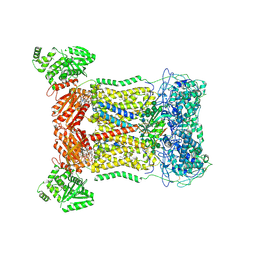 | | Cryo-EM structure of human DUOX1-DUOXA1 in low-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual oxidase 1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
7D3F
 
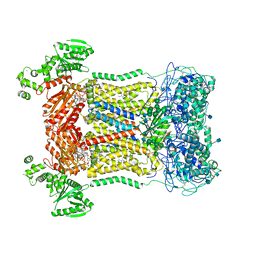 | | Cryo-EM structure of human DUOX1-DUOXA1 in high-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dual oxidase 1, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
7DXD
 
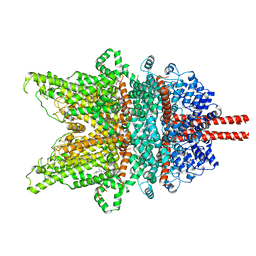 | | Structure of TRPC3 at 3.9 angstrom in 1340 nM free calcium state | | Descriptor: | CALCIUM ION, CHOLESTEROL HEMISUCCINATE, Short transient receptor potential channel 3, ... | | Authors: | Chen, L, Guo, W. | | Deposit date: | 2021-01-18 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism of human TRPC3 and TRPC6 channel regulation by their intracellular calcium-binding sites.
Neuron, 110, 2022
|
|
7VVH
 
 | |
7VUO
 
 | |
7VVD
 
 | |
7VLB
 
 | |
7D9R
 
 | | Structure of huamn soluble guanylate cyclase in the riociguat and NO-bound state | | Descriptor: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, MAGNESIUM ION, ... | | Authors: | Chen, L, Liu, R, Kang, Y. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
7D9U
 
 | | Structure of human soluble guanylate cyclase in the cinciguat-bound activated state | | Descriptor: | 4-({(4-carboxybutyl)[2-(2-{[4-(2-phenylethyl)benzyl]oxy}phenyl)ethyl]amino}methyl)benzoic acid, Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, ... | | Authors: | Chen, L, Liu, R, Kang, Y. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
7D9S
 
 | | Structure of huamn soluble guanylate cyclase in the YC1 and NO-bound state | | Descriptor: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, MAGNESIUM ION, ... | | Authors: | Chen, L, Liu, R, Kang, Y. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
7D9T
 
 | | Structure of human soluble guanylate cyclase in the cinciguat-bound inactive state | | Descriptor: | 4-({(4-carboxybutyl)[2-(2-{[4-(2-phenylethyl)benzyl]oxy}phenyl)ethyl]amino}methyl)benzoic acid, Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1 | | Authors: | Chen, L, Liu, R, Kang, Y. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
7E16
 
 | |
3K1Q
 
 | | Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics | | Descriptor: | Core protein VP6, Outer capsid VP5, Outer capsid VP7, ... | | Authors: | Cheng, L.P, Zhu, J, Hiu, W.H, Zhang, X.K, Honig, B, Fang, Q, Zhou, Z.H. | | Deposit date: | 2009-09-28 | | Release date: | 2010-03-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Backbone Model of an Aquareovirus Virion by Cryo-Electron Microscopy and Bioinformatics
J.Mol.Biol., 397, 2010
|
|
7BR3
 
 | | Crystal structure of the protein 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl dodecanoate, 4-[[(1R)-2-[5-(2-fluoranyl-3-methoxy-phenyl)-3-[[2-fluoranyl-6-(trifluoromethyl)phenyl]methyl]-4-methyl-2,6-bis(oxidanylidene)pyrimidin-1-yl]-1-phenyl-ethyl]amino]butanoic acid, ... | | Authors: | Cheng, L, Shao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the human gonadotropin-releasing hormone receptor GnRH1R reveals an unusual ligand binding mode.
Nat Commun, 11, 2020
|
|
3IZ3
 
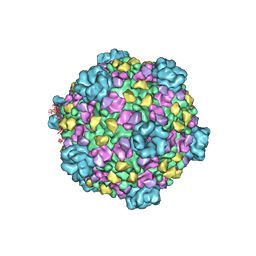 | | CryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Structural protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Cheng, L, Sun, J, Zhang, K, Mou, Z, Huang, X, Ji, G, Sun, F, Zhang, J, Zhu, P. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
