1IEH
 
 | | SOLUTION STRUCTURE OF A SOLUBLE SINGLE-DOMAIN ANTIBODY WITH HYDROPHOBIC RESIDUES TYPICAL OF A VL/VH INTERFACE | | Descriptor: | BRUC.D4.4 | | Authors: | Vranken, W, Tolkatchev, D, Xu, P, Tanha, J, Chen, Z, Narang, S, Ni, F. | | Deposit date: | 2001-04-09 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a llama single-domain antibody with hydrophobic residues typical of the VH/VL interface.
Biochemistry, 41, 2002
|
|
1WVE
 
 | | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon its Binding to the Cytochrome Subunit | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-cresol dehydrogenase [hydroxylating] cytochrome c subunit, 4-cresol dehydrogenase [hydroxylating] flavoprotein subunit, ... | | Authors: | Cunane, L.M, Chen, Z.-W, McIntire, W.S, Mathews, F.S. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon Its Binding to the Cytochrome Subunit
Biochemistry, 44, 2005
|
|
1WVF
 
 | | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon its Binding to the Cytochrome Subunit | | Descriptor: | 4-cresol dehydrogenase [hydroxylating] flavoprotein subunit, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Cunane, L.M, Chen, Z.-W, McIntire, W.S, Mathews, F.S. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon Its Binding to the Cytochrome Subunit
Biochemistry, 44, 2005
|
|
5D0N
 
 | | Crystal structure of maize PDRP bound with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Pyruvate, ... | | Authors: | Jiang, L, Chen, Z. | | Deposit date: | 2015-08-03 | | Release date: | 2016-02-24 | | Last modified: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Reversible Phosphorylation by Maize Pyruvate Orthophosphate Dikinase Regulatory Protein
Plant Physiol., 170, 2016
|
|
6NT5
 
 | | Cryo-EM structure of full-length human STING in the apo state | | Descriptor: | Stimulator of interferon protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
7X1N
 
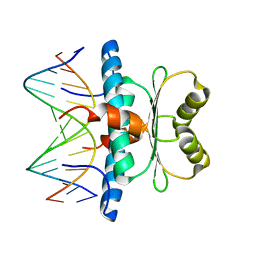 | | Crystal structure of MEF2D-MRE complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*G)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), Myocyte enhancer factor 2D/deleted in azoospermia associated protein 1 fusion protein | | Authors: | Zhang, H, Zhang, M, Wang, Q.Q, Chen, Z, Chen, S.J, Meng, G. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.315 Å) | | Cite: | Functional, structural, and molecular characterizations of the leukemogenic driver MEF2D-HNRNPUL1 fusion.
Blood, 140, 2022
|
|
6BJR
 
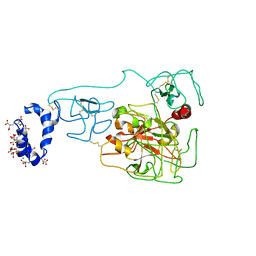 | | Crystal structure of prothrombin mutant S101C/A470C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Prothrombin, ... | | Authors: | Chinnaraj, M, Chen, Z, Pelc, L, Grese, Z, Bystranowska, D, Di Cera, E, Pozzi, N. | | Deposit date: | 2017-11-06 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structure of prothrombin in the closed form reveals new details on the mechanism of activation.
Sci Rep, 8, 2018
|
|
6NT6
 
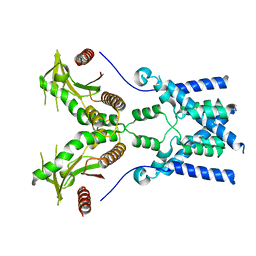 | | Cryo-EM structure of full-length chicken STING in the apo state | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
8JMM
 
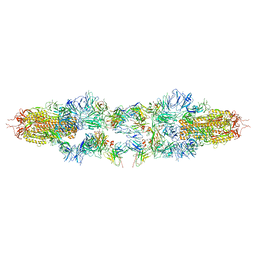 | | Structure of XBB spike protein (S) dimer-trimer in complex with bispecific antibody G7-Fc at 3.75 Angstroms resolution. | | Descriptor: | 7F3 Fv, GW01 Fv, Spike glycoprotein | | Authors: | Hao, A, Mao, Q, Chen, Z, Huang, J, Sun, L. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of XBB spike protein (S) dimer-trimer in complex with bispecific antibody G7-Fc at 3.75 Angstroms resolution.
To Be Published
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
6C2W
 
 | | Crystal structure of human prothrombin mutant S101C/A470C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Prothrombin, ... | | Authors: | Chinnaraj, M, Chen, Z, Pelc, L, Grese, Z, Bystranowska, D, Di Cera, E, Pozzi, N. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.12 Å) | | Cite: | Structure of prothrombin in the closed form reveals new details on the mechanism of activation.
Sci Rep, 8, 2018
|
|
5D1F
 
 | | Crystal structure of maize PDRP bound with AMP and Hg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Jiang, L, Chen, Z. | | Deposit date: | 2015-08-04 | | Release date: | 2016-02-24 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Reversible Phosphorylation by Maize Pyruvate Orthophosphate Dikinase Regulatory Protein
Plant Physiol., 170, 2016
|
|
7XA3
 
 | | Cryo-EM structure of the CCL2 bound CCR2-Gi complex | | Descriptor: | C-C motif chemokine 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
6NT9
 
 | | Cryo-EM structure of the complex between human TBK1 and chicken STING | | Descriptor: | Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of STING binding with and phosphorylation by TBK1.
Nature, 567, 2019
|
|
7X9Y
 
 | | Cryo-EM structure of the apo CCR3-Gi complex | | Descriptor: | C-C chemokine receptor type 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
6NT8
 
 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound tetrameric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
7VLA
 
 | | Cryo-EM structure of the CCL15(27-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(27-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL8
 
 | | Cryo-EM structure of the Apo CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL9
 
 | | Cryo-EM structure of the CCL15(26-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(26-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VGJ
 
 | | Cryo-EM structure of the human P4-type flippase ATP8B1-CDC50A in the auto-inhibited E2Pi-PS state | | Descriptor: | Cell cycle control protein 50A, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Phospholipid-transporting ATPase IC | | Authors: | Chen, M.T, Chen, Y, Chen, Z.P, Zhou, C.Z, Hou, W.T, Chen, Y. | | Deposit date: | 2021-09-16 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural insights into the activation of autoinhibited human lipid flippase ATP8B1 upon substrate binding.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6NT7
 
 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound dimeric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
8PNR
 
 | | Structure of human KCTD15 BTB domain mutant G88D crystal form 1 | | Descriptor: | BTB/POZ domain-containing protein KCTD15 | | Authors: | Cruz Walma, D.A, Cros, J, Bradshaw, W, Richardson, W, Chen, Z, Chalk, R, Wilkie, A, Bullock, A.N. | | Deposit date: | 2023-06-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | BTB domain mutations perturbing KCTD15 oligomerisation cause a distinctive frontonasal dysplasia syndrome.
J Med Genet, 61, 2024
|
|
1T31
 
 | | A Dual Inhibitor of the Leukocyte Proteases Cathepsin G and Chymase with Therapeutic Efficacy in Animals Models of Inflammation | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[3-({METHYL[1-(2-NAPHTHOYL)PIPERIDIN-4-YL]AMINO}CARBONYL)-2-NAPHTHYL]-1-(1-NAPHTHYL)-2-OXOETHYLPHOSPHONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | de Garavilla, L, Greco, M.N, Giardino, E.C, Wells, G.I, Haertlein, B.J, Kauffman, J.A, Corcoran, T.W, Derian, C.K, Eckardt, A.J, Abraham, W.M, Sukumar, N, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E, Andrade-Gordon, P, Damiano, B.P, Maryanoff, B.E, Pereira, P.J.B, Wang, Z.M, Rubin, H, Huber, R, Bode, W, Schechter, N.M, Strobl, S. | | Deposit date: | 2004-04-23 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel, potent dual inhibitor of the leukocyte proteases cathepsin G and chymase: molecular mechanisms and anti-inflammatory activity in vivo.
J.Biol.Chem., 280, 2005
|
|
7UQ2
 
 | | Vs.4 from T4 phage in complex with cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CALCIUM ION, Vs.4 | | Authors: | Jenson, J.M, Chen, Z.J. | | Deposit date: | 2022-04-18 | | Release date: | 2023-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ubiquitin-like conjugation by bacterial cGAS enhances anti-phage defence.
Nature, 616, 2023
|
|
7VJS
 
 | | Human AlkB homolog ALKBH6 in complex with Tris and Ni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, NICKEL (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
