6PCV
 
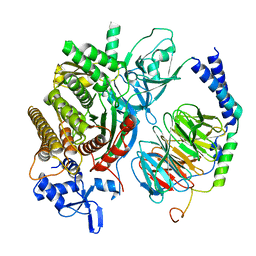 | | Single Particle Reconstruction of Phosphatidylinositol (3,4,5) trisphosphate-dependent Rac exchanger 1 bound to G protein beta gamma subunits | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Phosphatidylinositol (3,4,5) trisphosphate-dependent Rac exchanger 1 | | Authors: | Cash, J.N, Cianfrocco, M.A, Tesmer, J.J.G. | | Deposit date: | 2019-06-18 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-electron microscopy structure and analysis of the P-Rex1-G beta gamma signaling scaffold.
Sci Adv, 5, 2019
|
|
6BNM
 
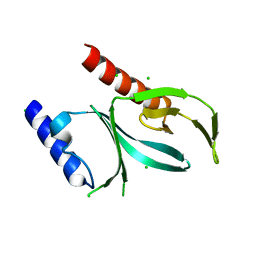 | | Crystal Structure of the P-Rex2 PH domain | | Descriptor: | CHLORIDE ION, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 2 protein | | Authors: | Cash, J.N, Sharma, P.V, Tesmer, J.J.G. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of the pleckstrin homology domain of the RhoGEF P-Rex2 and its regulation by PIP3.
J.Struct.Biol., 1, 2019
|
|
8TUA
 
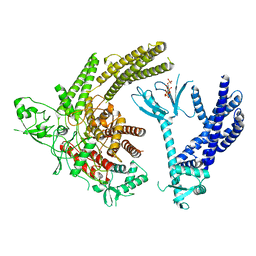 | | Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4) | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4)
Elife, 2024
|
|
3SEK
 
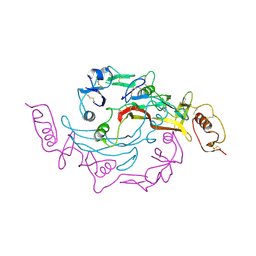 | | Crystal Structure of the Myostatin:Follistatin-like 3 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Follistatin-related protein 3, Growth/differentiation factor 8 | | Authors: | Cash, J.N, Thompson, T.B. | | Deposit date: | 2011-06-10 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of myostatinfollistatin-like 3: N-terminal domains of follistatin-type molecules exhibit alternate modes of binding.
J.Biol.Chem., 287, 2012
|
|
5FI1
 
 | | Crystal Structure of the P-Rex1 DH/PH tandem in complex with Cdc42 | | Descriptor: | Cell division control protein 42 homolog, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein,Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2015-12-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
5FI0
 
 | | Crystal Structure of the P-Rex1 DH/PH tandem in complex with Rac1 | | Descriptor: | PHOSPHATE ION, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein,Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2015-12-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.282 Å) | | Cite: | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
3HH2
 
 | |
5D3X
 
 | | Crystal Structure of the P-Rex1 PH domain with Inositol-(1,3,4,5)-Tetrakisphosphate Bound | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2015-08-06 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
5D3W
 
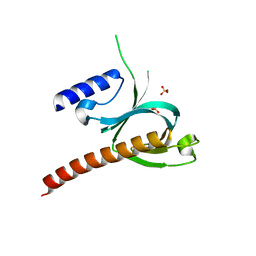 | | Crystal Structure of the P-Rex1 PH domain with Sulfate Bound | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein, SULFATE ION | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2015-08-06 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
5D27
 
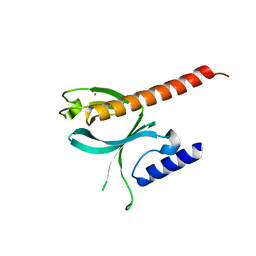 | | Crystal Structure of the P-Rex1 PH domain | | Descriptor: | NICKEL (II) ION, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2015-08-05 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
5D3Y
 
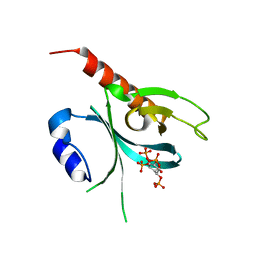 | | Crystal Structure of the P-Rex1 PH domain with Inositol-(1,3,4,5)-Tetrakisphosphate Bound | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2015-08-06 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
5D3V
 
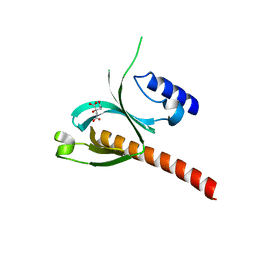 | | Crystal Structure of the P-Rex1 PH domain with Citrate Bound | | Descriptor: | CITRATE ANION, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2015-08-06 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
7KA3
 
 | | Aldolase, rabbit muscle (beam-tilt refinement x3) | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Kearns, S.K, Cash, J.N, Cianfrocco, M.A, Li, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High-resolution cryo-EM using beam-image shift at 200 keV.
Iucrj, 7, 2020
|
|
7KA2
 
 | | Aldolase, rabbit muscle (beam-tilt refinement x2) | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Kearns, S.K, Cash, J.N, Cianfrocco, M.A, Li, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution cryo-EM using beam-image shift at 200 keV.
Iucrj, 7, 2020
|
|
7KA4
 
 | | Aldolase, rabbit muscle (beam-tilt refinement x4) | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Kearns, S.K, Cash, J.N, Cianfrocco, M.A, Li, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution cryo-EM using beam-image shift at 200 keV.
Iucrj, 7, 2020
|
|
7K9X
 
 | |
7K9L
 
 | | Aldolase, rabbit muscle (no beam-tilt refinement) | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Cianfrocco, M.A, Kearns, S.E, Cash, J.N, Li, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | High-resolution cryo-EM using beam-image shift at 200 keV.
Iucrj, 7, 2020
|
|
6WJG
 
 | | PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma | | Descriptor: | DnaJ homolog subfamily B member 1, cAMP-dependent protein kinase catalytic subunit alpha fusion, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Lu, T.-W, Aoto, P.C, Weng, J.-H, Nielsen, C, Cash, J.N, Hall, J, Zhang, P, Simon, S.M, Cianfrocco, M.A, Taylor, S.S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural analyses of the PKA RII beta holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma.
Plos Biol., 18, 2020
|
|
6WJF
 
 | | PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma | | Descriptor: | DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha fusion, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Lu, T.-W, Aoto, P.C, Weng, J.-H, Nielsen, C, Cash, J.N, Hall, J, Zhang, P, Simon, S.M, Cianfrocco, M.A, Taylor, S.S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structural analyses of the PKA RII beta holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma.
Plos Biol., 18, 2020
|
|
