6I81
 
 | | Crystal Structure of the second bromodomain of BRD2 in complex with RT56 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2, ... | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of the second bromodomain of BRD2 in complex with RT56
To Be Published
|
|
6I7Y
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with RT56 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, [1-[4-[2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]ethanoylamino]phenyl]piperidin-4-yl]-trimethyl-azanium | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of the first bromodomain of BRD4 in complex with RT56
To Be Published
|
|
6I1S
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with FKBP12 and the inhibitor E6201 | | Descriptor: | (4~{S},5~{R},6~{Z},9~{S},10~{S},12~{E})-16-(ethylamino)-4,5-dimethyl-9,10,18-tris(oxidanyl)-3-oxabicyclo[12.4.0]octadeca-1(14),6,12,15,17-pentaene-2,8-dione, 1,2-ETHANEDIOL, Activin receptor type-1, ... | | Authors: | Williams, E.P, Pinkas, D.M, Fortin, J, Newman, J.A, Bradshaw, W.J, Mahajan, P, Kupinska, K, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-10-30 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Mutant ACVR1 Arrests Glial Cell Differentiation to Drive Tumorigenesis in Pediatric Gliomas.
Cancer Cell, 37, 2020
|
|
4XY8
 
 | | Crystal Structure of the bromodomain of BRD9 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 6-(5-bromo-2-methoxyphenyl)-9H-purin-2-amine, BROMIDE ION, Bromodomain-containing protein 9 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
4XY9
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-(5-bromo-2-methoxyphenyl)-9H-purin-2-amine, Bromodomain-containing protein 4 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
4XYA
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-(5-bromo-1-benzofuran-7-yl)-9H-purin-2-amine, Bromodomain-containing protein 4 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
6FAE
 
 | | The Sec7 domain of IQSEC2 (Brag1) in complex with the small GTPase Arf1 | | Descriptor: | 1,2-ETHANEDIOL, ADP-ribosylation factor 1, IQ motif and SEC7 domain-containing protein 2 | | Authors: | Gray, J, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P, von Delft, F. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Targeting the Small GTPase Superfamily through their Regulatory Proteins.
Angew.Chem.Int.Ed.Engl., 2019
|
|
6IBD
 
 | | The Phosphatase and C2 domains of Human SHIP1 | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Williams, E.P, Fernandez-Cid, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-11-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
6I5I
 
 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound 12h | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-[1-(phenylmethyl)pyrazol-4-yl]furo[3,2-b]pyridine, Dual specificity protein kinase CLK1 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
7NSG
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B | | Descriptor: | (+)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, (-)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S.M.M, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B
TO BE PUBLISHED
|
|
6I5H
 
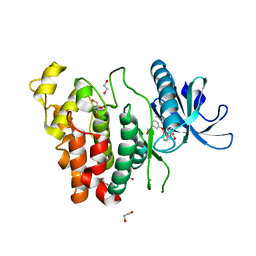 | | Crystal structure of CLK1 in complex with furanopyrimidin VN412 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-(3-phenoxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, ... | | Authors: | Schroeder, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
4UYI
 
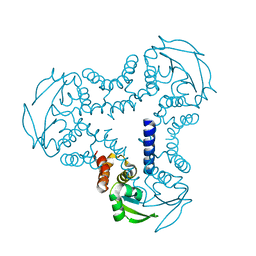 | | Crystal structure of the BTB domain of human SLX4 (BTBD12) | | Descriptor: | STRUCTURE-SPECIFIC ENDONUCLEASE SUBUNIT SLX4 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Strain-Damerell, C, Fairhead, M, Wang, D, Tallant, C, Cooper, C.D.O, Sorrell, F.J, Kopec, J, Chaikuad, A, Fitzpatrick, F, Pike, A.C.W, Hozjan, V, Ying, Z, Roos, A.K, Savitsky, P, Bradley, A, Nowak, R, Filippakopoulos, P, Krojer, T, Burgess-Brown, N.A, Marsden, B.D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-09-01 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the Btb Domain of Human Slx4 (Btbd12)
To be Published
|
|
4UUC
 
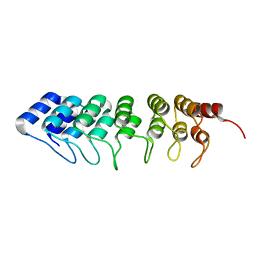 | | Crystal structure of human ASB11 ankyrin repeat domain | | Descriptor: | ANKYRIN REPEAT AND SOCS BOX PROTEIN 11 | | Authors: | Pinkas, D.M, Sanvitale, C, Kragh Nielsen, T, Guo, K, Sorrell, F, Berridge, G, Ayinampudi, V, Wang, D, Newman, J.A, Tallant, C, Chaikuad, A, Canning, P, Kopec, J, Krojer, T, Vollmar, M, Allerston, C.K, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Asb11 Ankyrin Repeat Domain
To be Published
|
|
7OR1
 
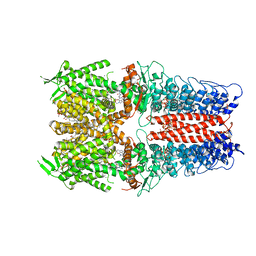 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
TO BE PUBLISHED
|
|
7OR0
 
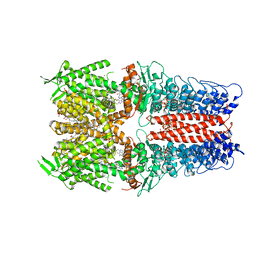 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
To Be Published
|
|
4XDK
 
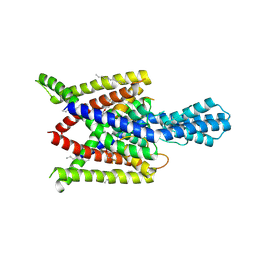 | | Crystal structure of human two pore domain potassium ion channel TREK2 (K2P10.1) in complex with norfluoxetine | | Descriptor: | (3R)-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, (3S)-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Mackenzie, A, Mukhopadhyay, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | K2P channel gating mechanisms revealed by structures of TREK-2 and a complex with Prozac.
Science, 347, 2015
|
|
4XDL
 
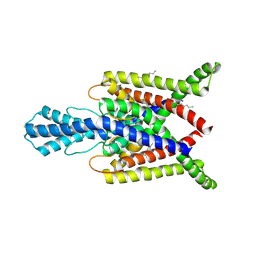 | | Crystal structure of human two pore domain potassium ion channel TREK2 (K2P10.1) in complex with a brominated fluoxetine derivative. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[2-bromanyl-4-(trifluoromethyl)phenoxy]-N-methyl-3-phenyl-propan-1-amine, CADMIUM ION, ... | | Authors: | Mackenzie, A, Pike, A.C.W, Dong, Y.Y, Mukhopadhyay, S, Ruda, G.F, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | K2P channel gating mechanisms revealed by structures of TREK-2 and a complex with Prozac.
Science, 347, 2015
|
|
4XDJ
 
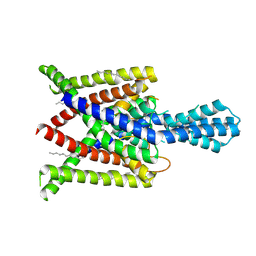 | | Crystal structure of human two pore domain potassium ion channel TREK2 (K2P10.1) in an alternate conformation (FORM 2) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, POTASSIUM CHANNEL SUBFAMILY K MEMBER 10, POTASSIUM ION, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Mackenzie, A, Mukhopadhyay, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | K2P channel gating mechanisms revealed by structures of TREK-2 and a complex with Prozac.
Science, 347, 2015
|
|
4WCI
 
 | | Crystal structure of the 1st SH3 domain from human CD2AP (CMS) in complex with a proline-rich peptide (aa 378-393) from human RIN3 | | Descriptor: | CD2-associated protein, Ras and Rab interactor 3, SULFATE ION | | Authors: | Rouka, E, Simister, P.C, Janning, M, Kirsch, K.H, Krojer, T, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-04 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).
J.Biol.Chem., 290, 2015
|
|
5C8H
 
 | | Crystal structure of ORC2 C-terminal domain | | Descriptor: | Origin recognition complex subunit 2, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Xu, C, Dong, A, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-25 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of ORC2 C-terminal domain
To Be Published
|
|
8F0W
 
 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956 | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)-3-fluorophenyl]butan-1-one, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-04 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956
to be published
|
|
7OZX
 
 | | Structure of human galactokinase 1 bound with azepan-1-yl(2,6-difluorophenyl)methanone | | Descriptor: | (azepan-1-yl)(2,6-difluorophenyl)methanone, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human galactokinase 1 bound with azepan-1-yl(2,6-difluorophenyl)methanone
To Be Published
|
|
8W3V
 
 | | Crystal structure of human WDR41 | | Descriptor: | WD repeat-containing protein 41 | | Authors: | Hutchinson, A, Dong, A, Li, Y, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human WDR41
To be published
|
|
5F5C
 
 | | Crystal Structure of human JMJD2D complexed with KDOPP7 | | Descriptor: | 1,2-ETHANEDIOL, 8-[[(phenylmethyl)amino]methyl]-1~{H}-pyrido[3,4-d]pyrimidin-4-one, Lysine-specific demethylase 4D, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F5A
 
 | | Crystal Structure of human JMJD2D complexed with KDOAM16 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(furan-2-ylmethylamino)methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4D, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
