4Y30
 
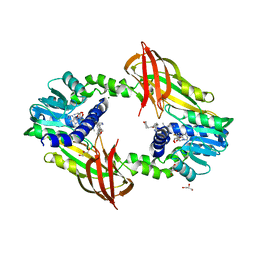 | | Crystal structure of human protein arginine methyltransferase PRMT6 bound to SAH and EPZ020411 | | Descriptor: | GLYCEROL, MAGNESIUM ION, N,N'-dimethyl-N-({3-[4-({trans-3-[2-(tetrahydro-2H-pyran-4-yl)ethoxy]cyclobutyl}oxy)phenyl]-1H-pyrazol-4-yl}methyl)ethane-1,2-diamine, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aryl Pyrazoles as Potent Inhibitors of Arginine Methyltransferases: Identification of the First PRMT6 Tool Compound.
Acs Med.Chem.Lett., 6, 2015
|
|
4XDO
 
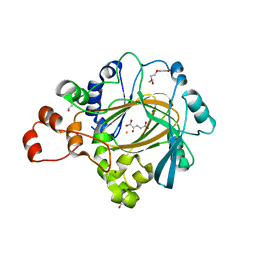 | | Crystal structure of human KDM4C catalytic domain with OGA | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Lysine-specific demethylase 4C, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A High-Throughput Mass Spectrometry Assay Coupled with Redox Activity Testing Reduces Artifacts and False Positives in Lysine Demethylase Screening.
J Biomol Screen, 20, 2015
|
|
4Y2H
 
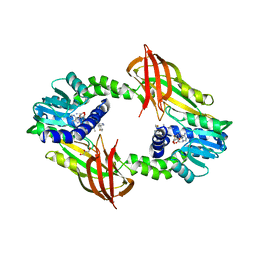 | |
8SZS
 
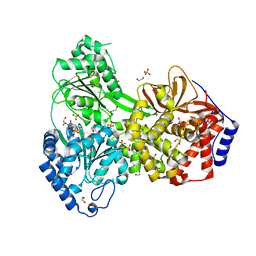 | | Cat DHX9 bound to GDP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-09-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Development of assays to support identification and characterization of modulators of DExH-box helicase DHX9.
Slas Discov, 28, 2023
|
|
8SZQ
 
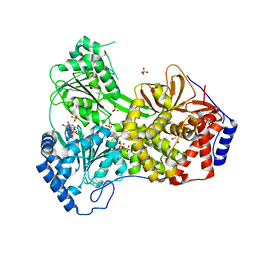 | | Cat DHX9 bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8SZP
 
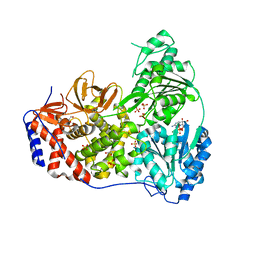 | | Human DHX9 bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase A, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8SZR
 
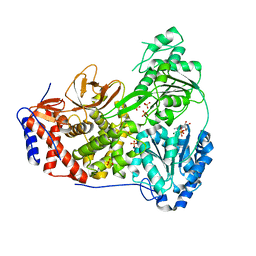 | | Dog DHX9 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RNA helicase, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4W5S
 
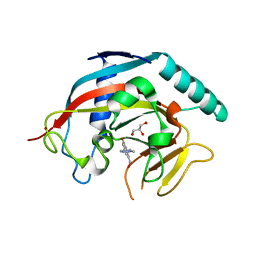 | | Tankyrase in complex with compound | | Descriptor: | 8-(hydroxymethyl)-2-[4-(1-methyl-1H-pyrazol-4-yl)phenyl]quinazolin-4(3H)-one, GLYCEROL, Tankyrase-1, ... | | Authors: | Johannes, J, Kazmirski, S.L, Boriack-Sjodin, P.A, Howard, T. | | Deposit date: | 2014-08-18 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyrimidinone nicotinamide mimetics as selective tankyrase and wnt pathway inhibitors suitable for in vivo pharmacology.
Acs Med.Chem.Lett., 6, 2015
|
|
4XDP
 
 | | Crystal structure of human KDM4C catalytic domain bound to tris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A High-Throughput Mass Spectrometry Assay Coupled with Redox Activity Testing Reduces Artifacts and False Positives in Lysine Demethylase Screening.
J Biomol Screen, 20, 2015
|
|
4UFZ
 
 | | Synthesis of Novel NAD Dependant DNA Ligase Inhibitors via Negishi Cross-Coupling: Development of SAR and Resistance Studies | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5,7-bis(azanyl)-2-tert-butyl-4-(1,3-thiazol-2-yl)pyrido[2,3-d]pyrimidine-6-carbonitrile, DNA LIGASE | | Authors: | Murphy-Benenato, K.E, Boriack-Sjodin, P.A, Martinez-Botella, G, Carcanague, D, Gingipali, L, Gowravaram, M, Harang, J, Hale, M, Ioannidis, G, Jahic, H, Johnstone, M, Kutschke, A, Laganas, V.A, Loch, J, Oguto, H, Patel, S.J. | | Deposit date: | 2015-03-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Negishi Cross-Coupling Enabled Synthesis of Novel Nad(+)-Dependent DNA Ligase Inhibitors and Sar Development.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4UCV
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-methoxy-2,3-dimethylquinoxalin-5-ol, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCT
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-amino-6-methyl-5-(propan-2-yloxy)-3H-[1,2,4]triazolo[1,5-a]pyrimidin-8-ium, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCO
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 7-amino-2-tert-butyl-4-(1H-pyrrol-2-yl)pyrido[2,3-d]pyrimidine-6-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCS
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5-amino-3-(furan-2-yl)-1H-1,2,4-triazole-1-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCU
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxyquinoline-2-carboxylic acid, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCR
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxy-2-methylquinoline-6-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
2B2X
 
 | | VLA1 RdeltaH I-domain complexed with a quadruple mutant of the AQC2 Fab | | Descriptor: | Antibody AQC2 Fab, Integrin alpha-1, MAGNESIUM ION | | Authors: | Clark, L.A, Boriack-Sjodin, P.A, Eldredge, J, Fitch, C, Friedman, B, Hanf, K.J, Jarpe, M, Liparoto, S.F, Li, Y, Lugovskoy, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Affinity enhancement of an in vivo matured therapeutic antibody using structure-based computational design
Protein Sci., 15, 2006
|
|
2ASK
 
 | | Structure of human Artemin | | Descriptor: | SULFATE ION, artemin | | Authors: | Silvian, L, Jin, P, Carmillo, P, Boriack-Sjodin, P.A, Pelletier, C, Rushe, M, Gong, B.J, Sah, D, Pepinsky, B, Rossomando, A. | | Deposit date: | 2005-08-23 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Artemin crystal structure reveals insights into heparan sulfate binding.
Biochemistry, 45, 2006
|
|
4KNR
 
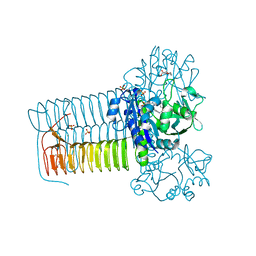 | | Hin GlmU bound to WG188 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, N-{4-[(2-benzyl-7-hydroxy-6-methoxyquinazolin-4-yl)amino]phenyl}benzamide, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-10 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
4KPZ
 
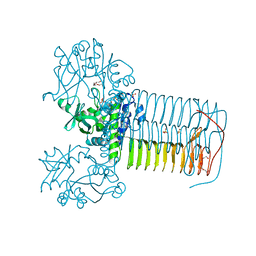 | | Hin GlmU bound to a small molecule fragment | | Descriptor: | 1-(3-nitrophenyl)dihydropyrimidine-2,4(1H,3H)-dione, Bifunctional protein GlmU, MAGNESIUM ION, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-14 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
4LH6
 
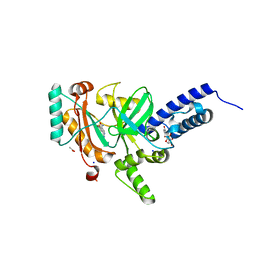 | | Crystal structure of a LigA inhibitor | | Descriptor: | 4-amino-2-bromothieno[3,2-c]pyridine-7-carboxamide, ACETATE ION, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ... | | Authors: | Benenato, K, Wang, H, Mcguire, H.M, Davis, H, Gao, N, Prince, D.B, Jahic, H, Stokes, S.S, Boriack-Sjodin, P.A. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification through structure-based methods of a bacterial NAD(+)-dependent DNA ligase inhibitor that avoids known resistance mutations.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4LP0
 
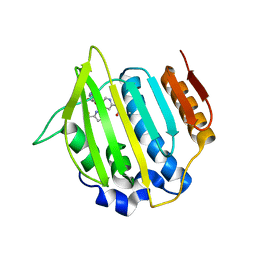 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid, Topoisomerase IV subunit B | | Authors: | Basarab, G.S, Manchester, J.I, Bist, S, Boriack-Sjodin, P.A, Dangel, B, Illingsworth, R, Uria-Nickelsen, M, Sherer, B.A, Sriram, S, Eakin, A.E. | | Deposit date: | 2013-07-14 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-to-Hit-to-Lead Discovery of a Novel Pyridylurea Scaffold of ATP Competitive Dual Targeting Type II Topoisomerase Inhibiting Antibacterial Agents.
J.Med.Chem., 56, 2013
|
|
4KNX
 
 | | Hin GlmU Bound to WG176 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-10 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
4KPX
 
 | | Hin GlmU bound to WG766 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, N-{4-[(4-hydroxy-3-nitrobenzoyl)amino]phenyl}pyridine-2-carboxamide, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-14 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
4KQL
 
 | | Hin GlmU bound to WG578 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, N-(4-{[3-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-5-methoxybenzoyl]amino}phenyl)pyridine-2-carboxamide, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-15 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
