1NU2
 
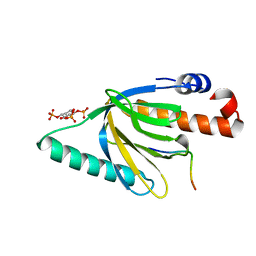 | | Crystal structure of the murine Disabled-1 (Dab1) PTB domain-ApoER2 peptide-PI-4,5P2 ternary complex | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Disabled homolog 1, peptide derived from murine Apolipoprotein E Receptor-2 | | Authors: | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | Deposit date: | 2003-01-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
1PB5
 
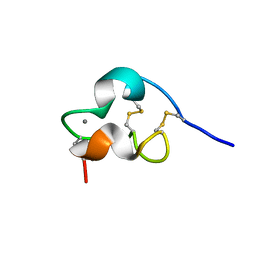 | | NMR Structure of a Prototype LNR Module from Human Notch1 | | Descriptor: | CALCIUM ION, Neurogenic locus notch homolog protein 1 | | Authors: | Vardar, D, North, C.L, Sanchez-Irizarry, C, Aster, J.C, Blacklow, S.C. | | Deposit date: | 2003-05-14 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of a Prototype Lin12-Notch Repeat Module from Human Notch1
Biochemistry, 42, 2003
|
|
1NTV
 
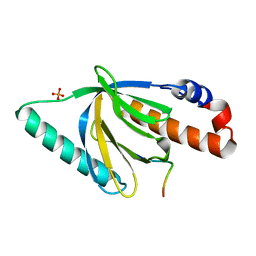 | | Crystal Structure of the Disabled-1 (Dab1) PTB domain-ApoER2 peptide complex | | Descriptor: | Apolipoprotein E Receptor-2 peptide, Disabled homolog 1, PHOSPHATE ION | | Authors: | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | Deposit date: | 2003-01-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
1Q68
 
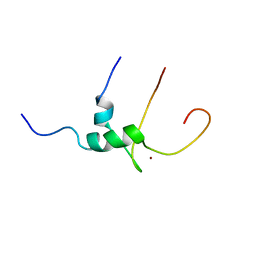 | | Solution structure of T-cell surface glycoprotein CD4 and Proto-oncogene tyrosine-protein kinase LCK fragments | | Descriptor: | Proto-oncogene tyrosine-protein kinase LCK, T-cell surface glycoprotein CD4, ZINC ION | | Authors: | Kim, P.W, Sun, Z.Y, Blacklow, S.C, Wagner, G, Eck, M.J. | | Deposit date: | 2003-08-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8.
Science, 301, 2003
|
|
1Q69
 
 | | Solution structure of T-cell surface glycoprotein CD8 alpha chain and Proto-oncogene tyrosine-protein kinase LCK fragments | | Descriptor: | Proto-oncogene tyrosine-protein kinase LCK, T-cell surface glycoprotein CD8 alpha chain, ZINC ION | | Authors: | Kim, P.W, Sun, Z.Y, Blacklow, S.C, Wagner, G, Eck, M.J. | | Deposit date: | 2003-08-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8.
Science, 301, 2003
|
|
5HQG
 
 | |
5IGO
 
 | |
5IGQ
 
 | |
5I6V
 
 | |
5TCX
 
 | | Crystal structure of human tetraspanin CD81 | | Descriptor: | CD81 antigen, CHOLESTEROL | | Authors: | Zimmerman, B, McMillan, B.J, Seegar, T.C.M, Kruse, A.C, Blacklow, S.C. | | Deposit date: | 2016-09-16 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket.
Cell, 167, 2016
|
|
5UX6
 
 | | Structure of Human POFUT1 in its apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GLYCEROL | | Authors: | Xu, X, McMillan, B, Blacklow, S.C. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations.
Glycobiology, 27, 2017
|
|
5VNY
 
 | |
5UXH
 
 | | Structure of Human POFUT1 in complex with GDP-fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE | | Authors: | Xu, X, McMillan, B, Blacklow, S.C. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations.
Glycobiology, 27, 2017
|
|
5VO5
 
 | |
5W55
 
 | |
6CIS
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG047 | | Descriptor: | 1,2-ETHANEDIOL, 11-cyclopentyl-5-methyl-2-({4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]-2-[(propan-2-yl)oxy]phenyl}amino)-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4, ... | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
6CIY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG069 | | Descriptor: | 1,2-ETHANEDIOL, 11-cyclobutyl-2-({2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4, ... | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
6CD4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG046 | | Descriptor: | 1,2-ETHANEDIOL, 2-({2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-11-(propan-2-yl)-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
6CD5
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor XMD17-26 | | Descriptor: | 1,2-ETHANEDIOL, 11-cyclopentyl-2-[[2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl-phenyl]amino]-5-methyl-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4, ... | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-08 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
6CJ2
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG056 | | Descriptor: | 2-{[3-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}-5-methyl-11-(propan-2-yl)-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
Acs Chem.Biol., 13, 2018
|
|
6CJ1
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG071 | | Descriptor: | 1,2-ETHANEDIOL, 11-[(2R)-butan-2-yl]-2-({2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, 11-[(2S)-butan-2-yl]-2-({2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, ... | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
2F8X
 
 | | Crystal structure of activated Notch, CSL and MAML on HES-1 promoter DNA sequence | | Descriptor: | 5'-D(*GP*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*AP*AP*A)-3', 5'-D(*TP*TP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*TP*AP*AP*C)-3', Mastermind-like protein 1, ... | | Authors: | Nam, Y, Sliz, P, Blacklow, S.C. | | Deposit date: | 2005-12-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes.
Cell(Cambridge,Mass.), 124, 2006
|
|
3NBN
 
 | |
2OO4
 
 | | Structure of LNR-HD (Negative Regulatory Region) from human Notch 2 | | Descriptor: | CALCIUM ION, GLYCEROL, Neurogenic locus notch homolog protein 2, ... | | Authors: | Gordon, W.R, Vardar-Ulu, D, Histen, G, Sanchez-Irizarry, C, Aster, J.C, Blacklow, S.C. | | Deposit date: | 2007-01-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis for autoinhibition of Notch
Nat.Struct.Mol.Biol., 14, 2007
|
|
3ETO
 
 | |
