1WM0
 
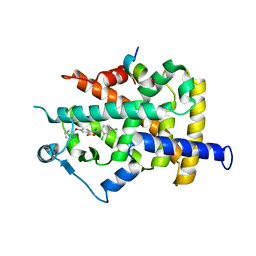 | | PPARgamma in complex with a 2-BABA compound | | Descriptor: | 14-mer from Nuclear receptor coactivator 2, 2-[(2,4-DICHLOROBENZOYL)AMINO]-5-(PYRIMIDIN-2-YLOXY)BENZOIC ACID, Peroxisome proliferator activated receptor gamma | | Authors: | Ostberg, T, Svensson, S, Selen, G, Uppenberg, J, Thor, M, Sundbom, M, Sydow-Backman, M, Gustavsson, A.L, Jendeberg, L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A new class of peroxisome proliferator-activated receptor agonists with a novel binding epitope shows antidiabetic effects
J.Biol.Chem., 279, 2004
|
|
6GN8
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 14-3-3 protein beta/alpha, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6HXS
 
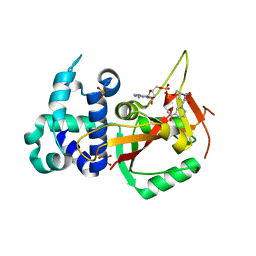 | | Human PARP16 (ARTD15) IN COMPLEX WITH CARBA-NAD | | Descriptor: | ADENOSINE, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Karlberg, T, Pinto, A.F, Thorsell, A.G, Schuler, H. | | Deposit date: | 2018-10-18 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Human PARP16 (ARTD15) IN COMPLEX WITH CARBA-NAD
To Be Published
|
|
6HXR
 
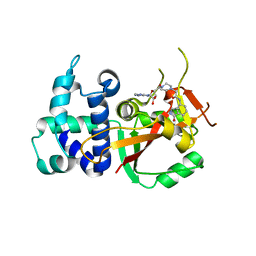 | | Human PARP16 (ARTD15) IN COMPLEX WITH EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Mono [ADP-ribose] polymerase PARP16 | | Authors: | Karlberg, T, Pinto, A.F, Thorsell, A.G, Schuler, H. | | Deposit date: | 2018-10-18 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Human PARP16 (ARTD15) IN COMPLEX WITH EB-47
To Be Published
|
|
6GN0
 
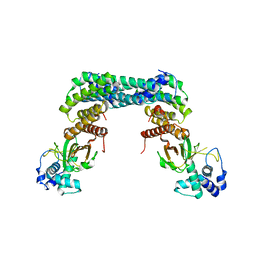 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, tetrameric crystal form | | Descriptor: | 14-3-3 protein beta/alpha, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-29 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GNN
 
 | | Exoenzyme T from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, tetrameric crystal form bound to STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme T | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GNJ
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form in complex with STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
4ABL
 
 | | HUMAN PARP14 (ARTD8, BAL2) - MACRO DOMAIN 3 | | Descriptor: | BROMIDE ION, POLY [ADP-RIBOSE] POLYMERASE 14 | | Authors: | Karlberg, T, Moche, M, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Recognition of Mono-Adp-Ribosylated Artd10 Substrates by Artd8 Macrodomains
Structure, 21, 2013
|
|
4ABK
 
 | | HUMAN PARP14 (ARTD8, BAL2) - MACRO DOMAIN 3 IN COMPLEX WITH ADENOSINE- 5-DIPHOSPHORIBOSE | | Descriptor: | POLY [ADP-RIBOSE] POLYMERASE 14, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Moche, M, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of Mono-Adp-Ribosylated Artd10 Substrates by Artd8 Macrodomains
Structure, 21, 2013
|
|
1L8X
 
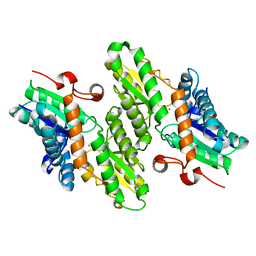 | | Crystal Structure of Ferrochelatase from the Yeast, Saccharomyces cerevisiae, with Cobalt(II) as the Substrate Ion | | Descriptor: | COBALT (II) ION, Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Metal Binding to Saccharomyces cerevisiae Ferrochelatase
Biochemistry, 41, 2002
|
|
3GOQ
 
 | |
1LBQ
 
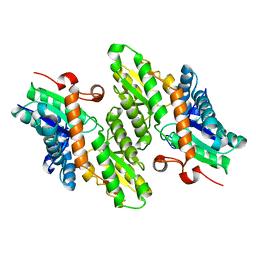 | | The crystal structure of Saccharomyces cerevisiae ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-04-04 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal binding to Saccharomyces cerevisiae ferrochelatase
Biochemistry, 41, 2002
|
|
4D86
 
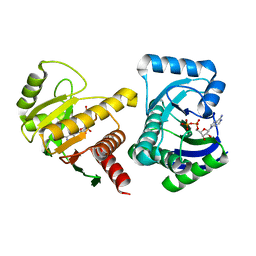 | | Human PARP14 (ARTD8, BAL2) - macro domains 1 and 2 in complex with adenosine-5-diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-01-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
1Z6C
 
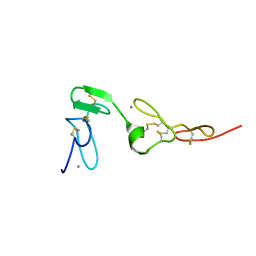 | | Solution structure of an EGF pair (EGF34) from vitamin K-dependent protein S | | Descriptor: | CALCIUM ION, Vitamin K-dependent protein S | | Authors: | Drakenberg, T, Ghasriani, H, Thulin, E, Thamlitz, A.M, Muranyi, A, Annila, A, Stenflo, J. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ca(2+)-Binding EGF3-4 Pair from Vitamin K-Dependent Protein S: Identification of an Unusual Fold in EGF3.
Biochemistry, 44, 2005
|
|
2Q2O
 
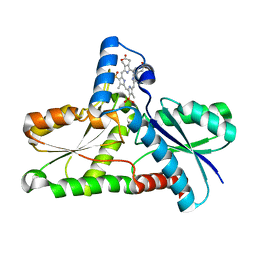 | | Crystal structure of H183C Bacillus subtilis ferrochelatase in complex with deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride | | Descriptor: | Ferrochelatase, MAGNESIUM ION, PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
2Q3J
 
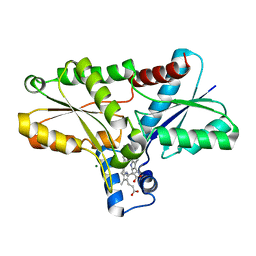 | | Crystal structure of the His183Ala variant of Bacillus subtilis ferrochelatase in complex with N-Methyl Mesoporphyrin | | Descriptor: | Ferrochelatase, MAGNESIUM ION, N-METHYL PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-30 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
2Q2N
 
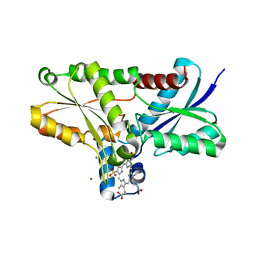 | | Crystal structure of Bacillus subtilis ferrochelatase in complex with deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride | | Descriptor: | Ferrochelatase, MAGNESIUM ION, PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-29 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
2GIL
 
 | | Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 resolution | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-6A | | Authors: | Bergbrede, T, Pylypenko, O, Rak, A, Alexandrov, K. | | Deposit date: | 2006-03-29 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 resolution
J.STRUCT.BIOL., 152, 2005
|
|
6EJP
 
 | | Yersinia YscU C-terminal fragment in complex with a synthetic compound | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Karlberg, T, Thorsell, A.G, Ho, O, Sunduru, N, Elofsson, M, Wolf-Watz, M, Schuler, H. | | Deposit date: | 2017-09-22 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Yersinia YscU C-terminal fragment in complex with a synthetic compound
To Be Published
|
|
3SMJ
 
 | | Human poly(ADP-ribose) polymerase 14 (Parp14/Artd8) - catalytic domain in complex with a pyrimidine-like inhibitor | | Descriptor: | 2-methyl-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one, GLYCEROL, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Graslund, S, Kouznetsova, E, Nordlund, P, Nyman, T, Thorsell, A.G, Tresaugues, L, Weigelt, J, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
3SE2
 
 | | Human poly(ADP-ribose) polymerase 14 (PARP14/ARTD8) - catalytic domain in complex with 6(5H)-phenanthridinone | | Descriptor: | 3-aminobenzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Graslund, S, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Thorsell, A.G, Tresaugues, L, Weigelt, J, Siponen, M.I, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
6FYM
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor ITK1 | | Descriptor: | 7,8-dimethyl-2-(pyrimidin-2-ylsulfanylmethyl)-3~{H}-quinazolin-4-one, NITRATE ION, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Kirby, I.T, Sreenivasan, R, Cohen, M.S, Schuler, H. | | Deposit date: | 2018-03-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Potent and Selective PARP11 Inhibitor Suggests Coupling between Cellular Localization and Catalytic Activity.
Cell Chem Biol, 25, 2018
|
|
3SMI
 
 | | Human poly(ADP-ribose) polymerase 14 (Parp14/Artd8) - catalytic domain in complex with a quinazoline inhibitor | | Descriptor: | 2-{[(3-amino-1H-1,2,4-triazol-5-yl)sulfanyl]methyl}-8-methylquinazolin-4(3H)-one, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Graslund, S, Kouznetsova, E, Nordlund, P, Nyman, T, Thorsell, A.G, Tresaugues, L, Weigelt, J, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
6FZM
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor ITK6 | | Descriptor: | 4-[(8-methyl-4-oxidanylidene-7-prop-1-ynyl-3~{H}-quinazolin-2-yl)methylsulfanyl]benzoic acid, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Kirby, I.T, Sreenivasan, R, Cohen, M.S, Schuler, H. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | A Potent and Selective PARP11 Inhibitor Suggests Coupling between Cellular Localization and Catalytic Activity.
Cell Chem Biol, 25, 2018
|
|
6G0W
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor MCD72 | | Descriptor: | 4-[3-[4-(4-fluorophenyl)piperidin-1-yl]carbonylphenoxy]benzamide, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Holechek, J, Lease, R, Ferraris, D, Schuler, H. | | Deposit date: | 2018-03-20 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Design, synthesis and evaluation of potent and selective inhibitors of mono-(ADP-ribosyl)transferases PARP10 and PARP14.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
