7JJU
 
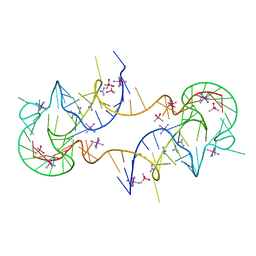 | | Crystal structure of en exoribonuclease-resistant RNA (xrRNA) from Potato leafroll virus (PLRV) | | Descriptor: | CACODYLATE ION, Guanidinium, IRIDIUM HEXAMMINE ION, ... | | Authors: | Steckelberg, A.-L, Vicens, Q, Auffinger, P, Costantino, D.C, Nix, J.C, Kieft, J.S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The crystal structure of a Polerovirus exoribonuclease-resistant RNA shows how diverse sequences are integrated into a conserved fold.
Rna, 26, 2020
|
|
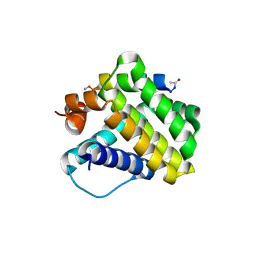
4HW4
 
 | |
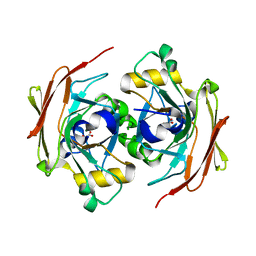
6FOG
 
 | |
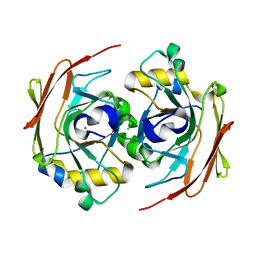
6FOH
 
 | |
2GBU
 
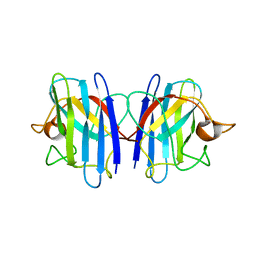 | | C6A/C111A/C57A/C146A apo CuZn Superoxide dismutase | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Hornberg, A, Logan, D.T, Marklund, S.L, Oliveberg, M. | | Deposit date: | 2006-03-11 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase
J.Mol.Biol., 365, 2007
|
|
7PGT
 
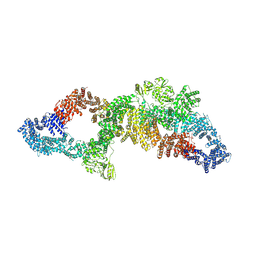 | | The structure of human neurofibromin isoform 2 in opened conformation. | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGS
 
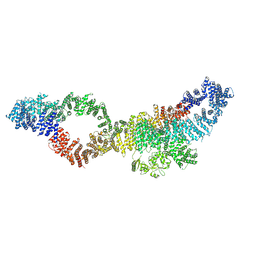 | | Consensus structure of human Neurofibromin isoform 2 | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGR
 
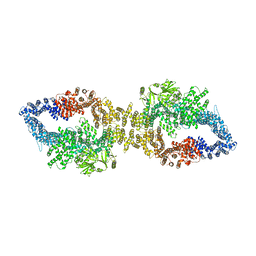 | | The structure of human neurofibromin isoform 2 in closed conformation | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGU
 
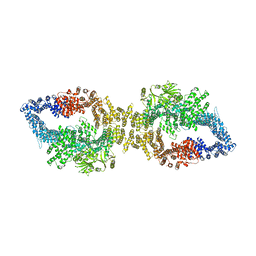 | | Autoinhibited structure of human neurofibromin isoform 2 stabilized by Zinc. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
6TAM
 
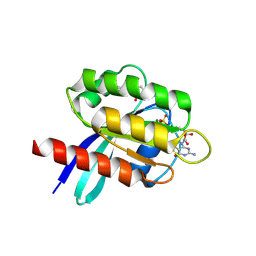 | |
8PAU
 
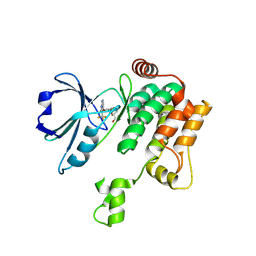 | | Crystal structure of MAP4K1 with a SMOL inhibitor | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1, [(5~{R})-2-[[3,5-bis(fluoranyl)-4-[[3-(trifluoromethyl)-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]oxy]phenyl]amino]-5-fluoranyl-4,6-dihydro-1,3-oxazin-5-yl]methanol | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
7PX3
 
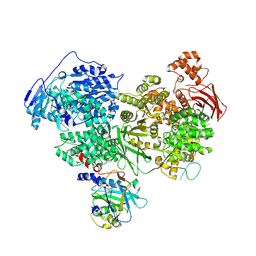 | | Structure of U5 snRNP assembly and recycling factor TSSC4 in complex with BRR2 and Jab1 domain of PRPF8 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Protein TSSC4, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Bergfort, A, Kuropka, B, Ilik, I.A, Freund, C, Aktas, T, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-10-07 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
2GBV
 
 | | C6A/C111A/C57A/C146A holo CuZn Superoxide dismutase | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Hornberg, A, Logan, D.T, Marklund, S.L, Oliveberg, M. | | Deposit date: | 2006-03-11 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase
J.Mol.Biol., 365, 2007
|
|
4KCC
 
 | | Crystal Structure of the NMDA Receptor GluN1 Ligand Binding Domain Apo State | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, PHOSPHATE ION | | Authors: | Berger, A.J, Lau, A.Y, Mayer, M.L. | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Conformational Analysis of NMDA Receptor GluN1, GluN2, and GluN3 Ligand-Binding Domains Reveals Subtype-Specific Characteristics.
Structure, 21, 2013
|
|
2GBT
 
 | | C6A/C111A CuZn Superoxide dismutase | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Hornberg, A, Logan, D.T, Marklund, S.L, Oliveberg, M. | | Deposit date: | 2006-03-11 | | Release date: | 2007-01-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase
J.Mol.Biol., 365, 2007
|
|
3TM0
 
 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa AMPPNP Butirosin A Complex | | Descriptor: | (2S)-4-amino-N-[(1R,2S,3R,4R,5S)-5-amino-4-[(2,6-diamino-2,6-dideoxy-alpha-D-glucopyranosyl)oxy]-2-hydroxy-3-(beta-D-xylofuranosyloxy)cyclohexyl]-2-hydroxybutanamide, Aminoglycoside 3'-phosphotransferase, MAGNESIUM ION, ... | | Authors: | Berghuis, A.M, Fong, D.H. | | Deposit date: | 2011-08-30 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of APH(3')-IIIa-mediated resistance to N1-substituted aminoglycoside antibiotics.
Antimicrob.Agents Chemother., 53, 2009
|
|
7OS1
 
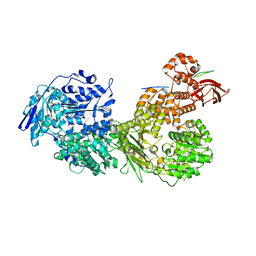 | | Cryo-EM structure of Brr2 in complex with Fbp21 | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase, WW domain-binding protein 4 | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
7OS2
 
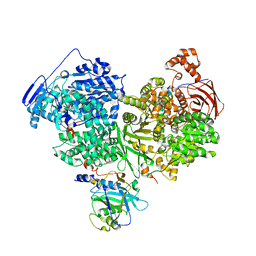 | | Cryo-EM structure of Brr2 in complex with Jab1/MPN and C9ORF78 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Telomere length and silencing protein 1 homolog, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
6SBU
 
 | | X-ray Structure of Human LDHA with an Allosteric Inhibitor (Compound 3) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-[[4-[(5-chloranylthiophen-2-yl)carbonylamino]-1,3-bis(oxidanylidene)isoindol-2-yl]methyl]benzoic acid, L-lactate dehydrogenase A chain | | Authors: | Friberg, A, Puetter, V, Nguyen, D, Rehwinkel, H. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Evidence for Isoform-Selective Allosteric Inhibition of Lactate Dehydrogenase A.
Acs Omega, 5, 2020
|
|
6SBV
 
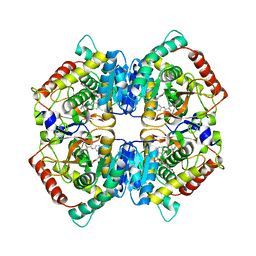 | | X-ray Structure of Human LDH-A with an Allosteric Inhibitor (Compound 7) | | Descriptor: | L-lactate dehydrogenase A chain, ~{N}-[3-[(7-nitrodibenzofuran-2-yl)sulfonylamino]phenyl]-1-oxidanyl-cyclopropane-1-carboxamide | | Authors: | Friberg, A, Puetter, V, Nguyen, D, Rehwinkel, H. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Evidence for Isoform-Selective Allosteric Inhibition of Lactate Dehydrogenase A.
Acs Omega, 5, 2020
|
|
7UGG
 
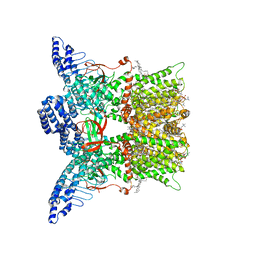 | | Cryo-EM structure of TRPV3 in complex with the anesthetic dyclonine | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Dyclonine, SODIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2022-03-24 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural mechanism of TRPV3 channel inhibition by the anesthetic dyclonine.
Nat Commun, 13, 2022
|
|
8PAW
 
 | | Crystal structure of MST1 with a MAP4K1 SMOL inhibitor | | Descriptor: | 1-[3,5-bis(fluoranyl)-4-[[3-(1-propan-2-ylpyrazol-3-yl)-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]oxy]phenyl]-3-(2-methoxyethyl)urea, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ASPARTIC ACID, ... | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
8PAS
 
 | | Crystal structure of MAP4K1 with a SMOL inhibitor | | Descriptor: | 4-[2,6-bis(fluoranyl)-4-(3-morpholin-4-ylpropylcarbamoylamino)phenoxy]-~{N}-[(4-methyl-1,2,5-oxadiazol-3-yl)methyl]-1~{H}-pyrrolo[2,3-b]pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
8PAV
 
 | | Crystal structure of MST1 with a MAP4K1 SMOL inhibitor | | Descriptor: | 1-[3,5-bis(fluoranyl)-4-[[3-(1,3-thiazol-5-yl)-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]oxy]phenyl]-3-(2-methoxyethyl)urea, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, ... | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
7PGP
 
 | |
