3P0P
 
 | | Human Tankyrase 2 - Catalytic PARP domain in complex with an inhibitor | | Descriptor: | 2-[4-(4-fluorophenyl)piperazin-1-yl]-6-methylpyrimidin-4(3H)-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Sehic, A, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
6G0S
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated SIRT7 peptide (K272ac/K275ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, NAD-dependent protein deacetylase sirtuin-7 | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
6TU9
 
 | | The ROR1 Pseudokinase Domain Bound To Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Inactive tyrosine-protein kinase transmembrane receptor ROR1 | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Niininen, W, Ungureanu, D, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
6G54
 
 | | Crystal structure of ERK2 covalently bound to SM1-71 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Mitogen-activated protein kinase 1, ... | | Authors: | Chaikuad, A, Suman, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
3K21
 
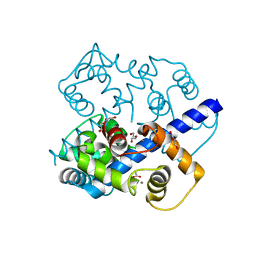 | | Crystal Structure of carboxy-terminus of PFC0420w. | | Descriptor: | ACETATE ION, CALCIUM ION, Calcium-dependent protein kinase 3, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Artz, J.D, Mackenzie, F, Cossar, D, Kozieradzki, I, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structures of parasitic CDPK domains point to a common mechanism of activation.
Proteins, 79, 2011
|
|
4A63
 
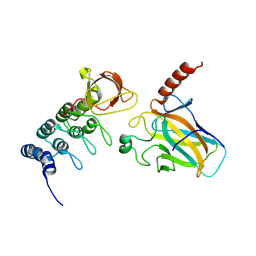 | | Crystal structure of the p73-ASPP2 complex at 2.6A resolution | | Descriptor: | ACETATE ION, APOPTOSIS STIMULATING OF P53 PROTEIN 2, TUMOUR PROTEIN 73, ... | | Authors: | Canning, P, Sharpe, T, Krojer, T, Savitsky, P, Cooper, C.D.O, Salah, E, Keates, T, Muniz, J, Vollmar, M, von Delft, F, Weigelt, J, Arrowsmith, C, Bountra, C, Edwards, A, Bullock, A.N. | | Deposit date: | 2011-10-31 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Basis for Aspp2 Recognition by the Tumor Suppressor P73.
J.Mol.Biol., 423, 2012
|
|
3MY0
 
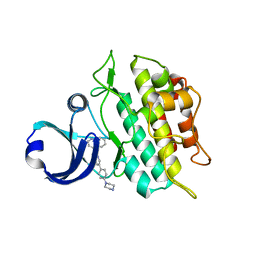 | | Crystal structure of the ACVRL1 (ALK1) kinase domain bound to LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Serine/threonine-protein kinase receptor R3 | | Authors: | Chaikuad, A, Alfano, I, Cooper, C, Mahajan, P, Daga, N, Sanvitale, C, Fedorov, O, Petrie, K, Savitsky, P, Gileadi, O, Sethi, R, Krojer, T, Muniz, J.R.C, Pike, A.C.W, Vollmar, M, Carpenter, C.P, Ugochukwu, E, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A small molecule targeting ALK1 prevents Notch cooperativity and inhibits functional angiogenesis.
Angiogenesis, 18, 2015
|
|
3K5K
 
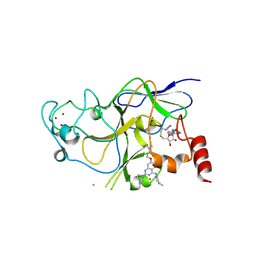 | | Discovery of a 2,4-Diamino-7-aminoalkoxy-quinazoline as a Potent Inhibitor of Histone Lysine Methyltransferase, G9a | | Descriptor: | 7-[3-(dimethylamino)propoxy]-6-methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinazolin-4-amine, CHLORIDE ION, Histone-lysine N-methyltransferase, ... | | Authors: | Dong, A, Wasney, G.A, Liu, F, Chen, X, Allali-Hassani, A, Senisterra, G, Chau, I, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Frye, S.V, Bochkarev, A, Brown, P.J, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a.
J.Med.Chem., 52, 2009
|
|
4AP2
 
 | | Crystal structure of the human KLHL11-Cul3 complex at 2.8A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CULLIN-3, ... | | Authors: | Canning, P, Cooper, C.D.O, Krojer, T, Filippakopoulos, P, Ayinampudi, V, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
6GI6
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 5-methyl core. | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-6-quinolin-5-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
3MET
 
 | | Crystal structure of SGF29 in complex with H3K4me2 | | Descriptor: | GLYCEROL, Histone H3, SAGA-associated factor 29 homolog, ... | | Authors: | Bian, C.B, Xu, C, Lam, R, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
3P0Q
 
 | | Human Tankyrase 2 - Catalytic PARP domain in complex with an inhibitor | | Descriptor: | N-[2-(4-chlorophenyl)ethyl]-6-methyl[1,2,4]triazolo[4,3-b]pyridazin-8-amine, SODIUM ION, SULFATE ION, ... | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Sehic, A, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
3DWY
 
 | | Crystal Structure of the Bromodomain of Human CREBBP | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Fedorov, O, Karim, R, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wickstroem, M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-23 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
6GIP
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 2, 5-dimethyl core. | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dimethyl-6-quinolin-4-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mathea, S, Chen, Z, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
6T1N
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
4A9C
 
 | | Crystal structure of human SHIP2 in complex with biphenyl 2,3',4,5',6- pentakisphosphate | | Descriptor: | BIPHENYL 2,3',4,5',6-PENTAKISPHOSPHATE, PHOSPHATIDYLINOSITOL-3,4,5-TRISPHOSPHATE 5-PHOSPHATASE 2 | | Authors: | Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Graslund, S, Karlberg, T, Mills, S.J, Moche, M, Nyman, T, Persson, C, Potter, B.V.L, Schuler, H, Thorsell, A.G, Weigelt, J, Nordlund, P. | | Deposit date: | 2011-11-25 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Synthetic Polyphosphoinositide Headgroup Surrogate in Complex with Ship2 Provides a Rationale for Drug Discovery.
Acs Chem.Biol., 7, 2012
|
|
3GEY
 
 | | Crystal structure of human poly(ADP-ribose) polymerase 15, catalytic fragment in complex with an inhibitor Pj34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Poly [ADP-ribose] polymerase 15 | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
4RXJ
 
 | | crystal structure of WHSC1L1-PWWP2 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Dong, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-11 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histone and DNA binding ability studies of the NSD subfamily of PWWP domains.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
4AVP
 
 | | Crystal structure of the DNA-binding domain of human ETV1. | | Descriptor: | 1,2-ETHANEDIOL, ETS TRANSLOCATION VARIANT 1 | | Authors: | Allerston, C.K, Cooper, C.D.O, Krojer, T, Chaikuad, A, Filippakopoulos, P, Canning, P, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2012-05-28 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
6G0Q
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated GATA1 peptide (K312ac/K315ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Erythroid transcription factor | | Authors: | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
6T1I
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-ethanoylphenyl)-~{N}-[(6-methoxypyridin-3-yl)methyl]piperazine-1-carboxamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1O
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-iodanyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
3HM6
 
 | | Crystal structure of the cytoplasmic domain of human plexin B1 | | Descriptor: | Plexin-B1, UNKNOWN ATOM OR ION, Unknown peptide | | Authors: | Tong, Y, He, H, Shen, L, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structure and function of the intracellular region of the plexin-b1 transmembrane receptor.
J.Biol.Chem., 284, 2009
|
|
3L50
 
 | | The crystal structure of human Glia Maturation Factor, Gamma (GMFG) | | Descriptor: | CHLORIDE ION, Glia maturation factor gamma | | Authors: | Ugochukwu, E, Pilka, E, Krysztofinska, E, Hapka, E, Krojer, T, Muniz, J, Vollmar, M, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human Glia Maturation Factor, Gamma (GMFG)
To be Published
|
|
6G0O
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated ATRX peptide (K1030ac/K1033ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Transcriptional regulator ATRX | | Authors: | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
