5E8R
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | CHLORIDE ION, N-methyl-N-({4-[4-(propan-2-yloxy)phenyl]-1H-pyrrol-3-yl}methyl)ethane-1,2-diamine, Protein arginine N-methyltransferase 6, ... | | Authors: | DONG, A, ZENG, H, LIU, J, TEMPEL, W, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, JIN, J, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases.
Acs Chem.Biol., 11, 2016
|
|
3CYN
 
 | | The structure of human GPX8 | | Descriptor: | GLYCEROL, Probable glutathione peroxidase 8, SULFATE ION | | Authors: | Kavanagh, K.L, Johansson, C, Yue, W.W, Kochan, G, Pike, A.C.W, Murray, J, Roos, A.K, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of human GPX8
To be Published
|
|
5APA
 
 | | Crystal structure of human aspartate beta-hydroxylase isoform a | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, ASPARTYL/ASPARAGINYL BETA-HYDROXYLASE, ... | | Authors: | Krojer, T, Kochan, G, Pfeffer, I, McDonough, M.A, Pilka, E, Hozjan, V, Allerston, C, Muniz, J.R, Chaikuad, A, Gileadi, O, Kavanagh, K, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2015-09-15 | | Release date: | 2015-09-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Aspartate/asparagine-beta-hydroxylase crystal structures reveal an unexpected epidermal growth factor-like domain substrate disulfide pattern.
Nat Commun, 10, 2019
|
|
4QC1
 
 | | Crystal structure of human BAZ2B bromodomain in complex with an acetylated histone 3 peptide (H3K14ac) | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, SULFATE ION, ZINC ION, ... | | Authors: | Tallant, C, Jose, B, Picaud, S, Chaikuad, A, Filippakopoulos, P, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-05-09 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
6R7Z
 
 | | CryoEM structure of calcium-free human TMEM16K / Anoctamin 10 in detergent (closed form) | | Descriptor: | Anoctamin-10 | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.14 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6RV3
 
 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 1000493 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6EIX
 
 | | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with a 2-aminopyridine inhibitor K02288 | | Descriptor: | 1,2-ETHANEDIOL, 3-[6-amino-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenol, Activin receptor type-1 | | Authors: | Williams, E.P, Canning, P, Sanvitale, C.E, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2017-09-19 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with K02288
To be published
|
|
4D0N
 
 | | AKAP13 (AKAP-Lbc) RhoGEF domain in complex with RhoA | | Descriptor: | 1,2-ETHANEDIOL, A-KINASE ANCHOR PROTEIN 13, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Abdul Azeez, K.R, Shrestha, L, Krojer, T, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A.M, Knapp, S, Klussmann, E, Elkins, J.M. | | Deposit date: | 2014-04-29 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Rhoa : Akap-Lbc Dh-Ph Domain Complex.
Biochem.J., 464, 2014
|
|
6RV4
 
 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 2341237 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, POTASSIUM ION, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6EQJ
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) Tyr195pIPhe mutant, apo form | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1 | | Authors: | Bailey, H.J, Kopec, J, Bilyard, M.K, Bezerra, G.A, Seo Lee, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Davis, B.G, Yue, W.W. | | Deposit date: | 2017-10-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Palladium-mediated enzyme activation suggests multiphase initiation of glycogenesis.
Nature, 563, 2018
|
|
6EKK
 
 | | Crystal structure of GEF domain of DENND 1A in complex with Rab GTPase Rab35-GDP bound state. | | Descriptor: | 1,2-ETHANEDIOL, DENN domain-containing protein 1A, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Srikannathasan, V, Szykowska, A, Tallant, C, Strain-Damerell, C, Kopec, J, Kupinska, K, Mukhopadhyay, S, Gavin, M, Wang, D, Chalk, R, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Huber, K. | | Deposit date: | 2017-09-26 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of DENND1A-RAB35 complex with GDP bound state.
To be published
|
|
6RV2
 
 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
5C7J
 
 | | CRYSTAL STRUCTURE OF NEDD4 WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
6S3Q
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with TFB-TBOA | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3)
TO BE PUBLISHED
|
|
4QTB
 
 | | Structure of human ERK1 in complex with SCH772984 revealing a novel inhibitor-induced binding pocket | | Descriptor: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Keates, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics.
Nat.Chem.Biol., 10, 2014
|
|
4QTE
 
 | | Structure of ERK2 in complex with VTX-11e, 4-{2-[(2-CHLORO-4-FLUOROPHENYL)AMINO]-5-METHYLPYRIMIDIN-4-YL}-N-[(1S)-1-(3-CHLOROPHENYL)-2-HYDROXYETHYL]-1H-PYRROLE-2-CARBOXAMIDE | | Descriptor: | 1,2-ETHANEDIOL, 4-{2-[(2-chloro-4-fluorophenyl)amino]-5-methylpyrimidin-4-yl}-N-[(1S)-1-(3-chlorophenyl)-2-hydroxyethyl]-1H-pyrrole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics.
Nat.Chem.Biol., 10, 2014
|
|
4LOQ
 
 | | Structural basis of autoactivation of p38 alpha induced by TAB1 (Tetragonal crystal form with bound sulphate) | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, Mitogen-activated protein kinase 14, ... | | Authors: | Chaikuad, A, DeNicola, G.F, Yue, W.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Marber, M.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Mechanism and consequence of the autoactivation of p38 alpha mitogen-activated protein kinase promoted by TAB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4QTA
 
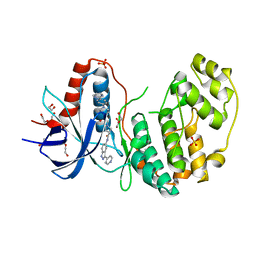 | | Structure of human ERK2 in complex with SCH772984 revealing a novel inhibitor-induced binding pocket | | Descriptor: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, 1,2-ETHANEDIOL, Mitogen-activated protein kinase 1, ... | | Authors: | Chaikuad, A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics.
Nat.Chem.Biol., 10, 2014
|
|
4QTC
 
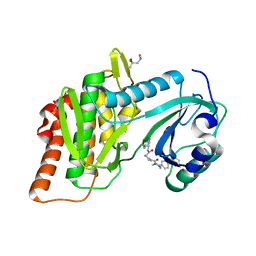 | | Structure of human haspin (GSG2) in complex with SCH772984 revealing the first type-I binding mode | | Descriptor: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, ... | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics.
Nat.Chem.Biol., 10, 2014
|
|
5A7Q
 
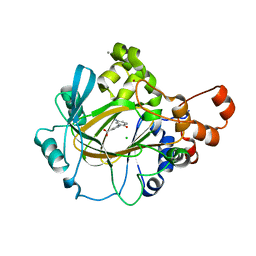 | | Crystal structure of human JMJD2A in complex with compound 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-(5-azanyl-2-oxidanyl-phenyl)pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Dixon-Clarke, S, MacKenzie, A, Nowak, R, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5A7N
 
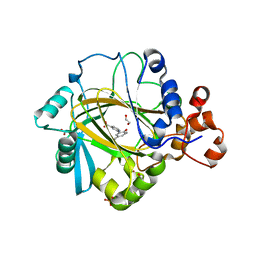 | | Crystal structure of human JMJD2A in complex with compound 43 | | Descriptor: | 1,2-ETHANEDIOL, 2-(5-cyano-2-oxidanyl-phenyl)pyridine-4-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Williams, E, Riesebos, E, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5AHR
 
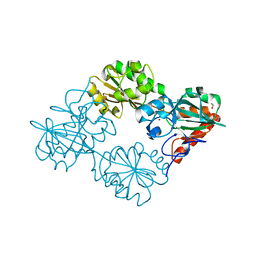 | | Crystal structure of human DNA cross-link repair 1A, crystal form B | | Descriptor: | 1,2-ETHANEDIOL, DNA CROSS-LINK REPAIR 1A PROTEIN, ZINC ION | | Authors: | Allerston, C.K, Newman, J.A, Vollmar, M, Goubin, S, Forese, D.S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2015-02-06 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Structures of the Snm1A and Snm1B/Apollo Nuclease Domains Reveal a Potential Basis for Their Distinct DNA Processing Activities.
Nucleic Acids Res., 43, 2015
|
|
6R7Y
 
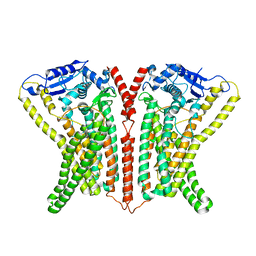 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (low Ca2+, closed form) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6R65
 
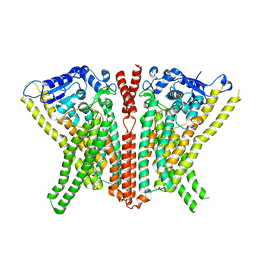 | | Crystal Structure of human TMEM16K / Anoctamin 10 (Form 2) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6FT9
 
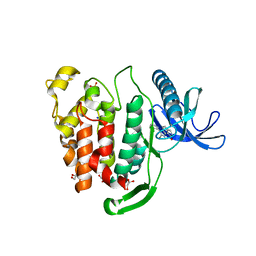 | | Crystal structure of CLK1 in complex with inhibitor 16 | | Descriptor: | 2-bromanyl-3-phenyl-1~{H}-pyrrolo[3,4-g]indol-8-one, BROMIDE ION, Dual specificity protein kinase CLK1, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
