2A5J
 
 | | Crystal Structure of Human RAB2B | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-2B | | Authors: | Dong, A, Wang, J, Shen, Y, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of human RAB2B
To be Published
|
|
3ZYW
 
 | | Crystal structure of the first glutaredoxin domain of human glutaredoxin 3 (GLRX3) | | Descriptor: | 1,2-ETHANEDIOL, GLUTAREDOXIN-3 | | Authors: | Vollmar, M, Johansson, C, Cocking, R, Krojer, T, Muniz, J.R.C, Kavanagh, K.L, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2011-08-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of the First Glutaredoxin Domain of Human Glutaredoxin 3 (Glrx3)
To be Published
|
|
2ABY
 
 | | Solution structure of TA0743 from Thermoplasma acidophilum | | Descriptor: | hypothetical protein TA0743 | | Authors: | Kim, B, Jung, J, Hong, E, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2005-07-18 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the conserved novel-fold protein TA0743 from Thermoplasma acidophilum.
Proteins, 62, 2006
|
|
2A14
 
 | | Crystal Structure of Human Indolethylamine N-methyltransferase with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, indolethylamine N-methyltransferase | | Authors: | Dong, A, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-17 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Human Indolethylamine
N-methyltransferase in complex with SAH.
To be Published
|
|
3ZYQ
 
 | | Crystal structure of the tandem VHS and FYVE domains of Hepatocyte growth factor-regulated tyrosine kinase substrate (HGS-Hrs) at 1.48 A resolution | | Descriptor: | 1,2-ETHANEDIOL, HEPATOCYTE GROWTH FACTOR-REGULATED TYROSINE KINASE SUBSTRATE, SULFATE ION, ... | | Authors: | Muniz, J.R.C, Ayinampudi, V, Shrestha, L, Krojer, T, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bullock, A. | | Deposit date: | 2011-08-24 | | Release date: | 2011-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of the Tandem Vhs and Fyve Domains of Hepatocyte Growth Factor-Regulated Tyrosine Kinase Substrate (Hgs-Hrs) at 1.48 A Resolution
To be Published
|
|
2B4Y
 
 | | Crystal Structure of Human Sirtuin homolog 5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-5, ... | | Authors: | Min, J.R, Antoshenko, T, Dong, A, Schuetz, A, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Sirtuin homolog 5 in complex with NAD
To be Published
|
|
4A27
 
 | | Crystal structure of human synaptic vesicle membrane protein VAT-1 homolog-like protein | | Descriptor: | 1,2-ETHANEDIOL, SYNAPTIC VESICLE MEMBRANE PROTEIN VAT-1 HOMOLOG-LIKE | | Authors: | Vollmar, M, Shafqat, N, Muniz, J.R.C, Krojer, T, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Yue, W.W, Oppermann, U. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Synaptic Vesicle Membrane Protein Vat-1 Homolog-Like Protein
To be Published
|
|
4A9Z
 
 | | CRYSTAL STRUCTURE OF HUMAN P63 TETRAMERIZATION DOMAIN | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, TUMOR PROTEIN 63 | | Authors: | Muniz, J.R.C, Coutandin, D, Salah, E, Chaikuad, A, Vollmar, M, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, von Delft, F, Knapp, S. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of Human P63 Tetramerization Domain
To be Published
|
|
8BI5
 
 | | Crystal structure of human Choline Kinase A in complex with UNC0737 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-6-methoxy-~{N}-methyl-~{N}-(1-propan-2-ylpiperidin-4-yl)-7-(3-pyrrolidin-1-ylpropoxy)quinazolin-4-amine, CHLORIDE ION, ... | | Authors: | Diaz-Saez, L, Ward, J, Kennedy, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human Choline Kinase A in complex with UNC0737
To Be Published
|
|
8BI6
 
 | | Crystal structure of human Choline Kinase A in complex with UNC0638 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-6-methoxy-N-[1-(1-methylethyl)piperidin-4-yl]-7-(3-pyrrolidin-1-ylpropoxy)quinazolin-4-amine, CHLORIDE ION, ... | | Authors: | Diaz-Saez, L, Ward, J, Kennedy, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human Choline Kinase A in complex with UNC0638
To Be Published
|
|
8D41
 
 | | Crystal structure of the human COPB2 WD-domain in complex with OICR-6254 | | Descriptor: | (1R,2R,3S,4S)-3-[4-(4-fluorophenyl)piperazine-1-carbonyl]bicyclo[2.2.1]heptane-2-carboxylic acid, Coatomer subunit beta', GLYCEROL, ... | | Authors: | Zeng, H, Saraon, P, Dong, A, Hutchinson, A, Seitova, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human COPB2 WD-domain in complex with OICR-6254
To Be Published
|
|
8D30
 
 | | Crystal structure of the human COPB2 WD-domains | | Descriptor: | 1,2-ETHANEDIOL, Coatomer subunit beta' | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-31 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human COPB2 WD-domains
To Be Published
|
|
7TUJ
 
 | | NMR solution structure of the phosphorylated MUS81-binding region from human SLX4 | | Descriptor: | Structure-specific endonuclease subunit SLX4 | | Authors: | Payliss, B.J, Reichheld, S.E, Lemak, A, Arrowsmith, C.H, Sharpe, S, Wyatt, H.D.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-09 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of the DNA repair scaffold SLX4 drives folding of the SAP domain and activation of the MUS81-EME1 endonuclease.
Cell Rep, 41, 2022
|
|
6AU2
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4H,6H-[1,2,4]triazolo[4,3-a][4,1]benzoxazepine, BETA-MERCAPTOETHANOL, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-10-11 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification and characterization of the first fragment hits for SETDB1 Tudor domain.
Bioorg.Med.Chem., 27, 2019
|
|
6AU3
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, N-{[2-(3,5-dimethyl-4H-1,2,4-triazol-4-yl)phenyl]methyl}acetamide, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments
to be published
|
|
6EI2
 
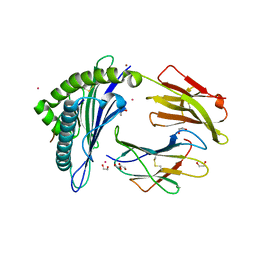 | | Crystal Structure of HLA-A68 presenting a C-terminally extended peptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Picaud, S, Guillaume, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gfeller, D, Filippakopoulos, P. | | Deposit date: | 2017-09-16 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of HLA-A68 presenting a C-terminally extended peptide
To Be Published
|
|
6EL8
 
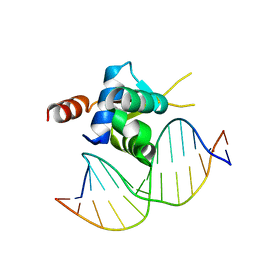 | | Crystal structure of the Forkhead domain of human FOXN1 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*GP*GP*CP*GP*TP*CP*TP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*AP*GP*AP*CP*GP*CP*CP*AP*CP*C)-3'), Forkhead box protein N1 | | Authors: | Newman, J.A, Aitkenhead, H.A, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of the Forkhead domain of human FOXN1 in complex with DNA
To be published
|
|
6EDF
 
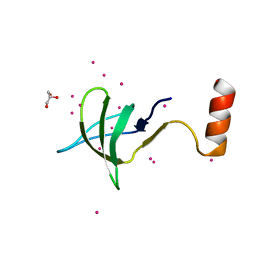 | | Fragment of a tyrosine-protein kinase | | Descriptor: | FYN, GLYCEROL, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Huang, H, Sochirca, I, Liu, K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Sidhu, S.S, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-08-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fragment of a tyrosine-protein kinase
To be Published
|
|
8EOM
 
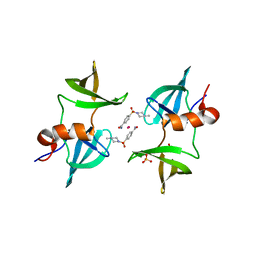 | | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-5973 | | Descriptor: | 4-(4-methylpiperazine-1-sulfonyl)benzamide, SULFATE ION, TP53-binding protein 1, ... | | Authors: | The, J, Hong, Z, Headey, S, Gunzburg, M, Doak, B, James, L.I, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-03 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-5973
to be published
|
|
8F0W
 
 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956 | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)-3-fluorophenyl]butan-1-one, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-04 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956
to be published
|
|
8FBG
 
 | | Crystal structure of NSD1 Mutant-Y1869C | | Descriptor: | CALCIUM ION, Histone-lysine N-methyltransferase, H3 lysine-36 specific, ... | | Authors: | Providokhina, K, Dong, A, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of NSD1
To Be Published
|
|
8FBH
 
 | | Crystal structure of NSD1 Mutant-T1927A | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Providokhina, K, Dong, A, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of NSD1
To Be Published
|
|
8F8E
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-8268 compound | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-3-(4-chloro-2-fluorophenyl)-1H-pyrazole-4-carboxamide, CITRIC ACID, DDB1- and CUL4-associated factor 1 | | Authors: | Kimani, S, Li, A, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-8268 compound
To be published
|
|
8G5E
 
 | | Crystal Structure of SETDB1 Tudor domain in complex with UNC6535 | | Descriptor: | Histone-lysine N-methyltransferase SETDB1, N~4~-[6-(dimethylamino)hexyl]-N~2~-[5-(dimethylamino)pentyl]-6,7-dimethoxyquinazoline-2,4-diamine, UNKNOWN ATOM OR ION | | Authors: | Beldar, S, Dong, A, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of SETDB1 Tudor domain in complex with UNC6535
To be published
|
|
8GCY
 
 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
