3KCI
 
 | | The third RLD domain of HERC2 | | Descriptor: | Probable E3 ubiquitin-protein ligase HERC2 | | Authors: | Walker, J.R, Qiu, L, Vesterberg, A, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Third RLD Domain of Herc2
To be Published
|
|
7N27
 
 | | Crystal Structure of chromodomain of CDYL in complex with inhibitor UNC6261 | | Descriptor: | Isoform 2 of Chromodomain Y-like protein, SODIUM ION, UNKNOWN ATOM OR ION, ... | | Authors: | Beldar, S, Dong, A, Loppnau, P, Min, J, Arrowsmith, C.H, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of chromodomain of CDYL in complex with inhibitor UNC6261
To Be Published
|
|
3V8D
 
 | | Crystal structure of human CYP7A1 in complex with 7-ketocholesterol | | Descriptor: | (3beta,8alpha,9beta)-3-hydroxycholest-5-en-7-one, Cholesterol 7-alpha-monooxygenase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Strushkevich, N, Tempel, W, MacKenzie, F, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Usanov, S.A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-22 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human CYP7A1 in complex with 7-ketocholesterol
To be Published
|
|
9MJG
 
 | | Crystal structure of HAT1 in complex with XS380871 | | Descriptor: | (4S,7R)-4-(3-ethoxy-4-hydroxy-5-nitrophenyl)-7-(4-fluorophenyl)-4,6,7,8-tetrahydroquinoline-2,5(1H,3H)-dione, Histone acetyltransferase type B catalytic subunit | | Authors: | Zeng, H, Li, F, Wang, X, Sun, J, Dong, A, Peng, H, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-12-15 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of HAT1 in complex with XS380871
To be published
|
|
9MK7
 
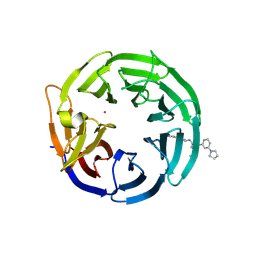 | | Crystal structure of the WDR domain of WDR91 in complex with DR3634 | | Descriptor: | (1R)-1-({(6M)-2-(methylamino)-6-[3-(pyrrolidin-1-yl)phenyl]pyrimidin-4-yl}amino)-2,3-dihydro-1H-indene-4-carbonitrile, UNKNOWN ATOM OR ION, WD repeat-containing protein 91 | | Authors: | Zeng, H, Ahmad, H, Dong, A, Seitova, A, Owen, D, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-12-16 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of the WDR domain of WDR91 in complex with DR3634
To be published
|
|
1HV2
 
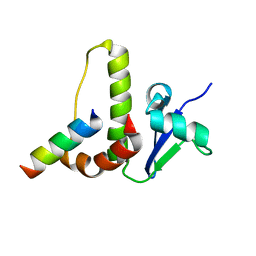 | | SOLUTION STRUCTURE OF YEAST ELONGIN C IN COMPLEX WITH A VON HIPPEL-LINDAU PEPTIDE | | Descriptor: | ELONGIN C, VON HIPPEL-LINDAU DISEASE TUMOR SUPPRESSOR | | Authors: | Botuyan, M.V, Mer, G, Yi, G.-S, Koth, C.M, Case, D.A, Edwards, A.M, Chazin, W.J, Arrowsmith, C.H. | | Deposit date: | 2001-01-05 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of yeast elongin C in complex with a von Hippel-Lindau peptide.
J.Mol.Biol., 312, 2001
|
|
1EP0
 
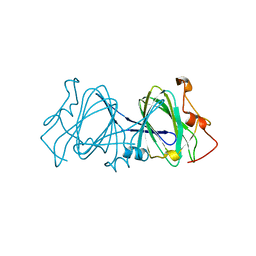 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-24 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
1ENW
 
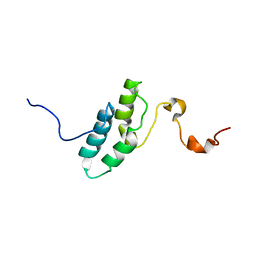 | |
9O0Y
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-[(2,3-dichlorophenyl)methyl]-3,5-difluoro-N-[(pyridin-3-yl)methyl]benzamide, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-04-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9O0X
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-({N-[(2,3-dichlorophenyl)methyl]-3,5-difluorobenzamido}methyl)-3-(methylamino)pyridine-2-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-04-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9O0W
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-({N-[(2,3-dichlorophenyl)methyl]-3,5-difluorobenzamido}methyl)pyridine-2-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-04-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9NIU
 
 | | Co-crystal structure of FABP7 in complex with PFO3TDA | | Descriptor: | 1,2-ETHANEDIOL, Fatty acid-binding protein, brain, ... | | Authors: | Yang, D, Liu, J, Zeng, H, Dong, A, Arrowsmith, C.H, Edwards, A.M, Peng, H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-02-26 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Co-crystal structure of FABP7 in complex with PFO3TDA
To be published
|
|
1EIJ
 
 | | NMR ENSEMBLE OF METHANOBACTERIUM THERMOAUTOTROPHICUM PROTEIN 1615 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1615 | | Authors: | Christendat, D, Booth, V, Gernstein, M, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-25 | | Release date: | 2000-11-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
3JWP
 
 | | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, TRIETHYLENE GLYCOL, Transcriptional regulatory protein sir2 homologue, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Lin, Y.H, MacKenzie, F, Senisterra, G, Allali-Hassanali, A, Vedadi, M, Ravichandran, M, Cossar, D, Kozieradzki, I, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Qiu, W, Brand, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-18 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP
TO BE PUBLISHED
|
|
6X9O
 
 | | High resolution cryoEM structure of huntingtin in complex with HAP40 | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin | | Authors: | Harding, R.J, Deme, J.C, Lea, S.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Huntingtin structure is orchestrated by HAP40 and shows a polyglutamine expansion-specific interaction with exon 1.
Commun Biol, 4, 2021
|
|
6XCG
 
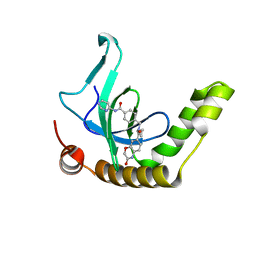 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound UNC6934 | | Descriptor: | Histone-lysine N-methyltransferase NSD2, N-cyclopropyl-3-oxo-N-({4-[(pyrimidin-4-yl)carbamoyl]phenyl}methyl)-3,4-dihydro-2H-1,4-benzoxazine-7-carboxamide, UNKNOWN ATOM OR ION | | Authors: | Zhou, M.Q, Dong, A, Ingerman, L.A, Hanley, R.P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
7YXX
 
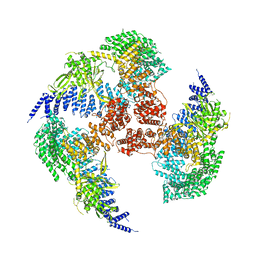 | |
7YXY
 
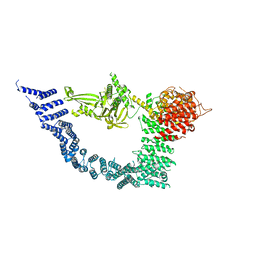 | |
3CON
 
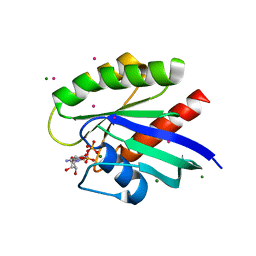 | | Crystal structure of the human NRAS GTPase bound with GDP | | Descriptor: | GTPase NRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Shen, L, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Crystal structure of the human NRAS GTPase bound with GDP.
To be Published
|
|
8W3V
 
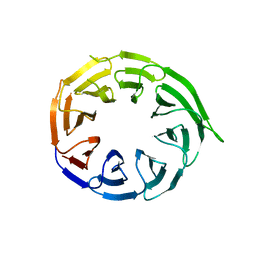 | | Crystal structure of human WDR41 | | Descriptor: | WD repeat-containing protein 41 | | Authors: | Hutchinson, A, Dong, A, Li, Y, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human WDR41
To be published
|
|
7HI8
 
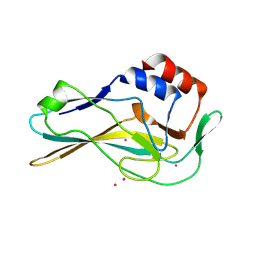 | | PanDDA analysis group deposition -- Crystal Structure of ground state model of human Brachyury | | Descriptor: | CADMIUM ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2024-09-20 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HI9
 
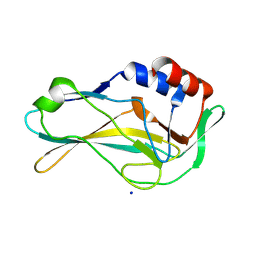 | | PanDDA analysis group deposition of ground-state model of human Brachyury G177D variant | | Descriptor: | SODIUM ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2024-09-20 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
7NSG
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B | | Descriptor: | (+)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, (-)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S.M.M, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B
TO BE PUBLISHED
|
|
7UGC
 
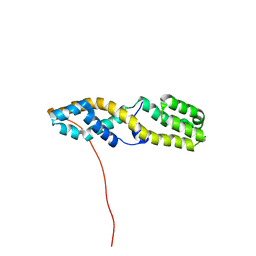 | | Structure of the N-terminal domain of ViaA | | Descriptor: | Protein ViaA | | Authors: | Lemak, A, Reichheld, S, Bhandari, V, Houliston, S, Arrowsmith, C.H, Sharpe, S, Houry, W.A. | | Deposit date: | 2022-03-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of ViaA
To Be Published
|
|
9GDK
 
 | | Jumonji domain-containing protein 1C with crystallization epitope mutations L2440Y:G2444H | | Descriptor: | Probable JmjC domain-containing histone demethylation protein 2C | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-05 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
