5C8Y
 
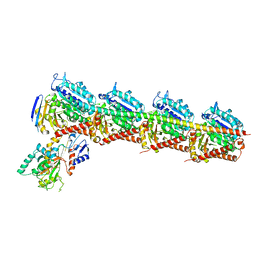 | | Crystal structure of T2R-TTL-Plinabulin complex | | 分子名称: | (3Z,6Z)-3-benzylidene-6-[(5-tert-butyl-1H-imidazol-4-yl)methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | 登録日 | 2015-06-26 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.594 Å) | | 主引用文献 | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
4RA2
 
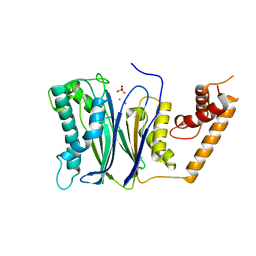 | | PP2Ca | | 分子名称: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | 著者 | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | 登録日 | 2014-09-09 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | The catalytic role of the M2 metal ion in PP2Ca
SCI REP, 2015
|
|
4WSR
 
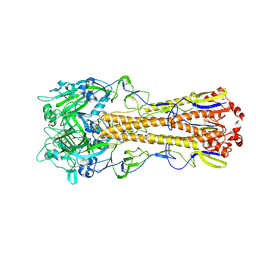 | | The crystal structure of hemagglutinin form A/chicken/New York/14677-13/1998 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | 著者 | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | 登録日 | 2014-10-28 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
5CA0
 
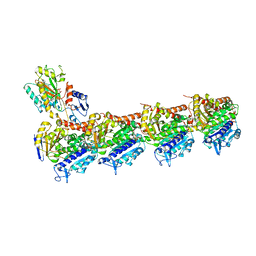 | | Crystal structure of T2R-TTL-Lexibulin complex | | 分子名称: | 1-ethyl-3-[2-methoxy-4-(5-methyl-4-{[(1S)-1-(pyridin-3-yl)butyl]amino}pyrimidin-2-yl)phenyl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | 登録日 | 2015-06-29 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
6X4Y
 
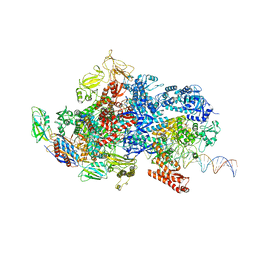 | | Mfd-bound E.coli RNA polymerase elongation complex - IV state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | 登録日 | 2020-05-24 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X50
 
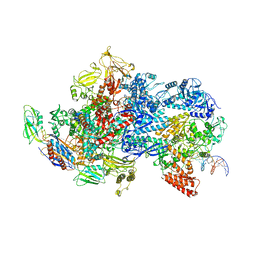 | | Mfd-bound E.coli RNA polymerase elongation complex - V state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Llewelyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | 登録日 | 2020-05-24 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
5C8C
 
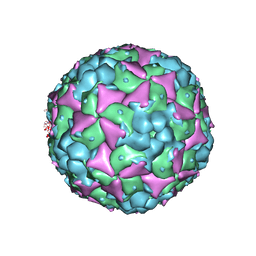 | | Crystal structure of recombinant coxsackievirus A16 capsid | | 分子名称: | CHLORIDE ION, POTASSIUM ION, STEARIC ACID, ... | | 著者 | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2015-06-25 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
6X26
 
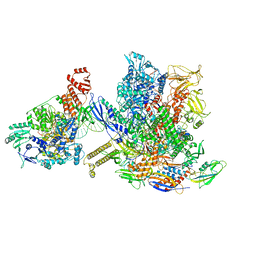 | | Mfd-bound E.coli RNA polymerase elongation complex - L1 state | | 分子名称: | DNA (64-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | 登録日 | 2020-05-20 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X43
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - II state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | 登録日 | 2020-05-22 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
5CB4
 
 | | Crystal structure of T2R-TTL-Tivantinib complex | | 分子名称: | (3R,4R)-3-(5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1-yl)-4-(1H-indol-3-yl)pyrrolidine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | 登録日 | 2015-06-30 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.193 Å) | | 主引用文献 | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
6K8N
 
 | | Crystal structure of the Sulfolobus solfataricus topoisomerase III | | 分子名称: | ZINC ION, topoisomerase III | | 著者 | Wang, H.Q, Zhang, J.H, Zheng, X, Zheng, Z.F, Dong, Y.H, Huang, L, Gong, Y. | | 登録日 | 2019-06-13 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of the Sulfolobus solfataricus topoisomerase III reveal that its C-terminal novel zinc finger part is a unique decatenation domain
To Be Published
|
|
6WR4
 
 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | 分子名称: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | 著者 | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | 登録日 | 2020-04-29 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
4WSS
 
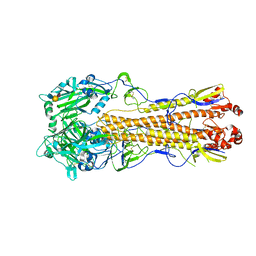 | | The crystal structure of hemagglutinin form A/chicken/New York/14677-13/1998 in complex with LSTa | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | 著者 | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | 登録日 | 2014-10-28 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
6WMQ
 
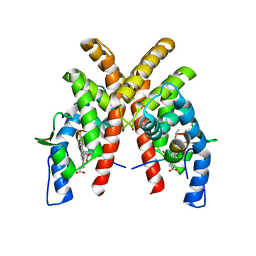 | |
6WMS
 
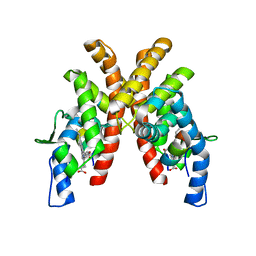 | |
6WDQ
 
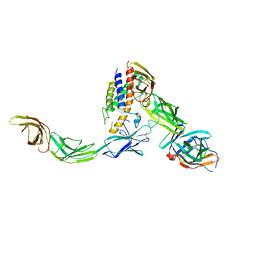 | | IL23/IL23R/IL12Rb1 signaling complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, ... | | 著者 | Jude, K.M, Ely, L.K, Glassman, C.R, Thomas, C, Spangler, J.B, Lupardus, P.J, Garcia, K.C. | | 登録日 | 2020-04-01 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
6GYU
 
 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-02 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5CTW
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(butanoylamino)thiophene-3-carboxamide, CHLORIDE ION, ... | | 著者 | Andersen, O.A, Barker, J, Hadfield, A.T, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CTX
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-phenyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, DNA gyrase subunit B, ... | | 著者 | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CPH
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (3E)-3-(pyridin-3-ylmethylidene)-1,3-dihydro-2H-indol-2-one, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit B, ... | | 著者 | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-21 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8FIV
 
 | |
8FIW
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor Jun10221 | | 分子名称: | 3C-like proteinase nsp5, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide, N-([1,1'-biphenyl]-4-yl)-N-[(1S)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide | | 著者 | Sacco, M, Wang, J, Chen, Y. | | 登録日 | 2022-12-16 | | 公開日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Exploring diverse reactive warheads for the design of SARS-CoV-2 main protease inhibitors.
Eur.J.Med.Chem., 259, 2023
|
|
3T9N
 
 | | Crystal structure of a membrane protein | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Small-conductance mechanosensitive channel | | 著者 | Yang, M, Zhang, X, Ge, J, Wang, J. | | 登録日 | 2011-08-03 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.456 Å) | | 主引用文献 | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5CTU
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(thiophen-2-yl)thieno[2,3-d]pyrimidin-4(1H)-one, CHLORIDE ION, ... | | 著者 | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8W8Q
 
 | | Cryo-EM structure of the GPR101-Gs complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Sun, J.P, Gao, N, Yu, X, Wang, G.P, Yang, F, Wang, J.Y, Yang, Z, Guan, Y. | | 登録日 | 2023-09-04 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
