2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | 分子名称: | hypothetical protein | | 著者 | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2006-02-09 | | 公開日 | 2006-02-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
7F09
 
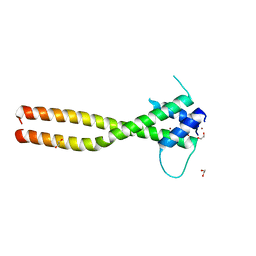 | | Crystal structure of the HLH-Lz domain of human TFE3 | | 分子名称: | 1,2-ETHANEDIOL, Transcription factor E3, ZINC ION | | 著者 | Yang, G, Li, P, Liu, Z, Wu, S, Zhuang, C, Qiao, H, Fang, P, Wang, J. | | 登録日 | 2021-06-03 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for the dimerization mechanism of human transcription factor E3.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
6WL2
 
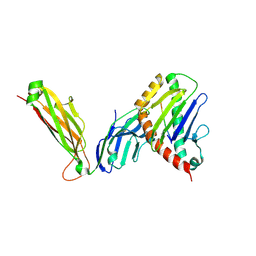 | | preTCRbeta-pMHC complex crystal structure | | 分子名称: | ARG-GLY-TYR-VAL-TYR-GLN-GLY-LEU, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | 著者 | Li, X, Mallis, R.J, Mizsei, R, Tan, K, Reinherz, E.L, Wang, J. | | 登録日 | 2020-04-18 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection.
Science, 371, 2021
|
|
7RHR
 
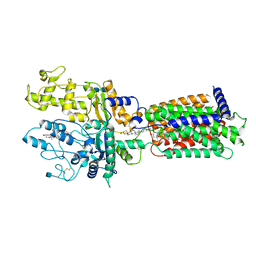 | | Cryo-EM structure of Xenopus Patched-1 in nanodisc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Huang, P, Lian, T, Jiang, J, Salic, A. | | 登録日 | 2021-07-18 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for catalyzed assembly of the Sonic hedgehog-Patched1 signaling complex.
Dev.Cell, 57, 2022
|
|
6WL4
 
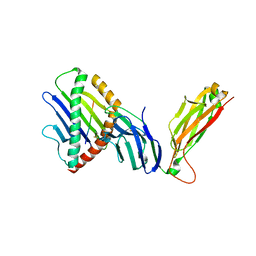 | | preTCRbeta-pMHC complex crystal structure | | 分子名称: | ARG-GLY-TYR-VAL-TYR-GLN-GLY-LEU, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | 著者 | Li, X, Mallis, R.J, Mizsei, R, Tan, K, Reinherz, E.L, Wang, J. | | 登録日 | 2020-04-18 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection.
Science, 371, 2021
|
|
4GB0
 
 | |
5J2Y
 
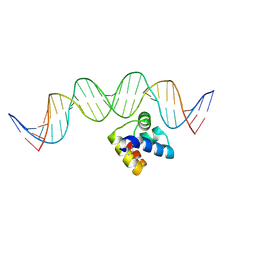 | | Molecular insight into the regulatory mechanism of the quorum-sensing repressor RsaL in Pseudomonas aeruginosa | | 分子名称: | DNA (26-MER), Regulatory protein | | 著者 | Zhao, J, Gan, J, Zhang, J, Kang, H, Kong, W, Zhu, M, Li, F, Song, Y, Qin, J, Liang, H. | | 登録日 | 2016-03-30 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing.
Nucleic Acids Res., 45, 2017
|
|
6IFY
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1 | | 分子名称: | CTR1, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
5A31
 
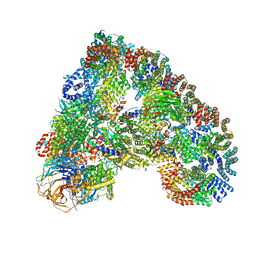 | | Structure of the human APC-Cdh1-Hsl1-UbcH10 complex. | | 分子名称: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | 著者 | Chang, L, Zhang, Z, Yang, J, Mclaughlin, S.H, Barford, D. | | 登録日 | 2015-05-26 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Atomic Structure of the Apc/C and its Mechanism of Protein Ubiquitination.
Nature, 522, 2015
|
|
5BOF
 
 | | Crystal Structure of Staphylococcus aureus Enolase | | 分子名称: | Enolase, MAGNESIUM ION, SULFATE ION | | 著者 | Wu, Y.F, Wang, C.L, Wu, M.H, Han, L, Zhang, X, Zang, J.Y. | | 登録日 | 2015-05-27 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3KCU
 
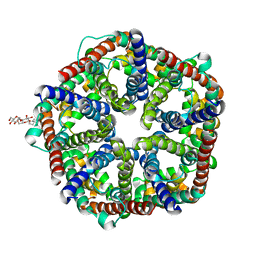 | | Structure of formate channel | | 分子名称: | 2-(6-(2-CYCLOHEXYLETHOXY)-TETRAHYDRO-4,5-DIHYDROXY-2(HYDROXYMETHYL)-2H-PYRAN-3-YLOXY)-TETRAHYDRO-6(HYDROXYMETHYL)-2H-PY RAN-3,4,5-TRIOL, Probable formate transporter 1 | | 著者 | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | 登録日 | 2009-10-22 | | 公開日 | 2009-12-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.243 Å) | | 主引用文献 | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
6JAU
 
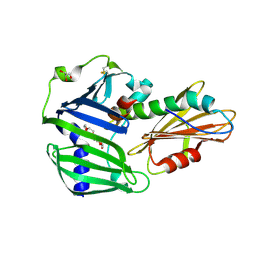 | | The complex structure of Pseudomonas aeruginosa MucA/MucB. | | 分子名称: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | 著者 | Li, T, He, L.H, Li, C.C, Liu, L, Peng, C.T, Shen, Y.L, Qin, X.F, Xiao, Q.J, Zhu, Y.B, Song, Y.J, Zhao, N.l, Zhao, C, Yang, J, Mu, X.Y, Huang, Q, Bao, R. | | 登録日 | 2019-01-25 | | 公開日 | 2020-01-29 | | 最終更新日 | 2020-08-19 | | 実験手法 | X-RAY DIFFRACTION (1.905 Å) | | 主引用文献 | Molecular basis of the lipid-induced MucA-MucB dissociation in Pseudomonas aeruginosa.
Commun Biol, 3, 2020
|
|
3IU3
 
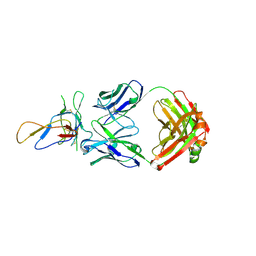 | | Crystal structure of the Fab fragment of therapeutic antibody Basiliximab in complex with IL-2Ra (CD25) ectodomain | | 分子名称: | Heavy chain of Fab fragment of Basiliximab, Interleukin-2 receptor alpha chain, Light chain of Fab fragment of Basiliximab, ... | | 著者 | Du, J, Yang, H, Wang, J, Ding, J. | | 登録日 | 2009-08-29 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for the blockage of IL-2 signaling by therapeutic antibody basiliximab
J.Immunol., 184, 2010
|
|
5TJA
 
 | | I-II linker of TRPML1 channel at pH 6 | | 分子名称: | Mucolipin-1 | | 著者 | Li, M, Zhang, W.K, Benvin, N.M, Zhou, X, Su, D, Li, H, Wang, S, Michailidis, I.E, Tong, L, Li, X, Yang, J. | | 登録日 | 2016-10-04 | | 公開日 | 2017-01-25 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5TJC
 
 | | I-II linker of TRPML1 channel at pH 7.5 | | 分子名称: | Mucolipin-1 | | 著者 | Li, M, Zhang, W.K, Benvin, N.M, Zhou, X, Su, D, Li, H, Wang, S, Michailidis, I.E, Tong, L, Li, X, Yang, J. | | 登録日 | 2016-10-04 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8WTE
 
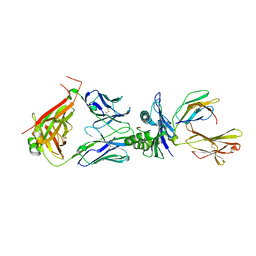 | | Crystal structure of TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | 分子名称: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen (Fragment), ... | | 著者 | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | 登録日 | 2023-10-18 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
6K2H
 
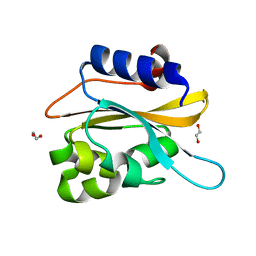 | | structural characterization of mutated NreA protein in nitrate binding site from staphylococcus aureus. | | 分子名称: | 1,2-ETHANEDIOL, NreA | | 著者 | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | 登録日 | 2019-05-14 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
7CRW
 
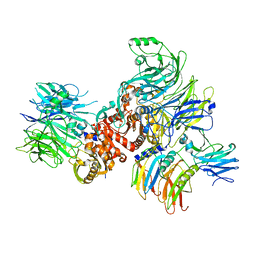 | | Cryo-EM structure of rNLRP1-rDPP9 complex | | 分子名称: | Dipeptidyl peptidase 9, NLR family protein 1 | | 著者 | Huang, M.H, Zhang, X.X, Wang, J, Chai, J.J. | | 登録日 | 2020-08-14 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Structural and biochemical mechanisms of NLRP1 inhibition by DPP9.
Nature, 592, 2021
|
|
7CRV
 
 | |
3IZ3
 
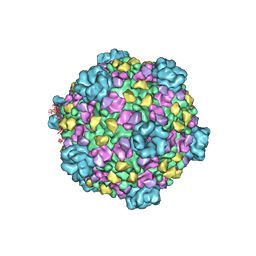 | | CryoEM structure of cytoplasmic polyhedrosis virus | | 分子名称: | Structural protein VP1, Structural protein VP3, Viral structural protein 5 | | 著者 | Cheng, L, Sun, J, Zhang, K, Mou, Z, Huang, X, Ji, G, Sun, F, Zhang, J, Zhu, P. | | 登録日 | 2010-09-14 | | 公開日 | 2011-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2IA0
 
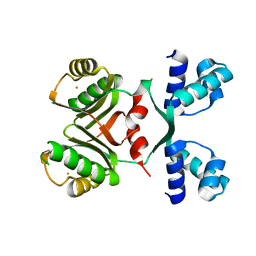 | | Transcriptional Regulatory Protein PF0864 From Pyrococcus Furiosus a Member of the ASNC Family (PF0864) | | 分子名称: | GOLD ION, Putative HTH-type transcriptional regulator PF0864 | | 著者 | Yang, H, Chang, J, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2006-09-06 | | 公開日 | 2006-10-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Crystal Structure of the Transcriptional Regulatory Protein PF0864: an Asnc Family member from Pyrococcus Furiosus
To be Published
|
|
8IHR
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | 分子名称: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | 分子名称: | Amidohydrolase family protein, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
6GYS
 
 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6WXK
 
 | | PHF23 PHD Domain Apo | | 分子名称: | PHD finger protein 23, ZINC ION | | 著者 | Vann, K.R, Zhang, J, Zhang, Y, Kutateladze, T. | | 登録日 | 2020-05-11 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Mechanistic insights into chromatin targeting by leukemic NUP98-PHF23 fusion.
Nat Commun, 11, 2020
|
|
