2OM9
 
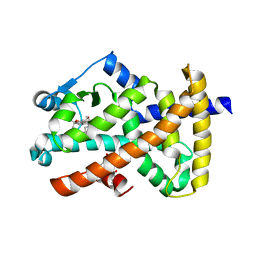 | | Ajulemic acid, a synthetic cannabinoid bound to PPAR gamma | | Descriptor: | (6AR,10AR)-3-(1,1-DIMETHYLHEPTYL)-1-HYDROXY-6,6-DIMETHYL-6A,7,10,10A-TETRAHYDRO-6H-BENZO[C]CHROMENE-9-CARBOXYLIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Ambrosio, A.L.B, Garratt, R.C. | | Deposit date: | 2007-01-21 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ajulemic Acid, a Synthetic Nonpsychoactive Cannabinoid Acid, Bound to the Ligand Binding Domain of the Human Peroxisome Proliferator-activated Receptor gamma
J.Biol.Chem., 282, 2007
|
|
8EC6
 
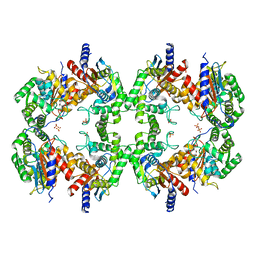 | | Cryo-EM structure of the Glutaminase C core filament (fGAC) | | Descriptor: | Isoform 2 of Glutaminase kidney isoform, mitochondrial, PHOSPHATE ION | | Authors: | Ambrosio, A.L, Dias, S.M, Quesnay, J.E, Portugal, R.V, Cassago, A, van Heel, M.G, Islam, Z, Rodrigues, C.T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of glutaminase activation through filamentation and the role of filaments in mitophagy protection.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3CQD
 
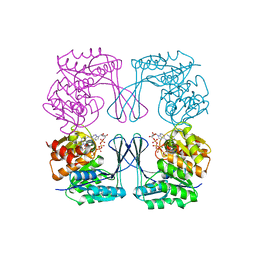 | | Structure of the tetrameric inhibited form of phosphofructokinase-2 from Escherichia coli | | Descriptor: | 6-phosphofructokinase isozyme 2, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Ambrosio, A.L, Cabrera, R, Caniuguir, A, Garratt, R.C, Babul, J. | | Deposit date: | 2008-04-02 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic structure of phosphofructokinase-2 from Escherichia coli in complex with two ATP molecules. Implications for substrate inhibition.
J.Mol.Biol., 383, 2008
|
|
3SS4
 
 | |
3SS5
 
 | |
3SS3
 
 | |
1S8I
 
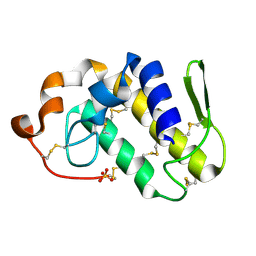 | | Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, second fatty acid free form | | Descriptor: | Phospholipase A2 homolog, SULFATE ION | | Authors: | Ambrosio, A.L.B, de Souza, D.H.F, Nonato, M.C, Selistre de Araujo, H.S, Ownby, C.L, Garratt, R.C. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | A Molecular Mechanism for Lys49-Phospholipase A2 Activity Based on Ligand-induced Conformational Change.
J.Biol.Chem., 280, 2005
|
|
1S8G
 
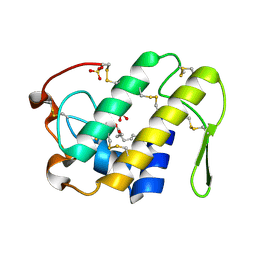 | | Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, fatty acid bound form | | Descriptor: | GLYCEROL, LAURIC ACID, Phospholipase A2 homolog, ... | | Authors: | Ambrosio, A.L.B, de Souza, D.H.F, Nonato, M.C, Selistre de Araujo, H.S, Ownby, C.L, Garratt, R.C. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Molecular Mechanism for Lys49-Phospholipase A2 Activity Based on Ligand-induced Conformational Change.
J.Biol.Chem., 280, 2005
|
|
1S8H
 
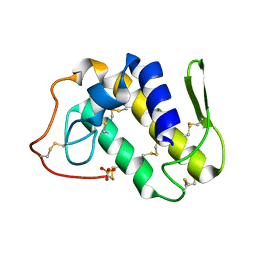 | | Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, first fatty acid free form | | Descriptor: | Phospholipase A2 homolog, SULFATE ION | | Authors: | Ambrosio, A.L.B, de Souza, D.H.F, Nonato, M.C, Selistre de Araujo, H.S, Ownby, C.L, Garratt, R.C. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Molecular Mechanism for Lys49-Phospholipase A2 Activity Based on Ligand-induced Conformational Change.
J.Biol.Chem., 280, 2005
|
|
4WN5
 
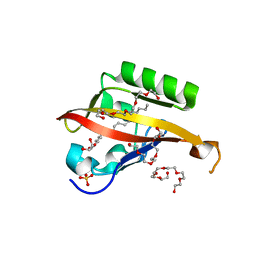 | | Crystal structure of the C-terminal Per-Arnt-Sim (PASb) of human HIF-3alpha9 bound to 18:1-1-monoacylglycerol | | Descriptor: | HEXAETHYLENE GLYCOL, Hypoxia-inducible factor 3-alpha, MONOVACCENIN, ... | | Authors: | Fala, A.M, Oliveira, J.F, Dias, S.M, Ambrosio, A.L. | | Deposit date: | 2014-10-10 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Unsaturated fatty acids as high-affinity ligands of the C-terminal Per-ARNT-Sim domain from the Hypoxia-inducible factor 3 alpha.
Sci Rep, 5, 2015
|
|
4JJM
 
 | | Structure of a cyclophilin from Citrus sinensis (CsCyp) in complex with cyclosporin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, cyclosporin A | | Authors: | Campos, B.M, Ambrosio, A.L.B, Souza, T.A.C.B, Barbosa, J.A.R.G, Benedetti, C.E. | | Deposit date: | 2013-03-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A redox 2-cys mechanism regulates the catalytic activity of divergent cyclophilins.
Plant Physiol., 162, 2013
|
|
4JKT
 
 | | Crystal structure of mouse Glutaminase C, BPTES-bound form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Fornezari, C, Ferreira, A.P.S, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2013-03-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Active Glutaminase C Self-assembles into a Supratetrameric Oligomer That Can Be Disrupted by an Allosteric Inhibitor.
J.Biol.Chem., 288, 2013
|
|
5F28
 
 | | Crystal structure of FAT domain of Focal Adhesion Kinase (FAK) bound to the transcription factor MEF2C | | Descriptor: | Focal adhesion kinase 1, MEF2C | | Authors: | Cardoso, A.C, Ambrosio, A.L.B, Dessen, A, Franchini, K.G. | | Deposit date: | 2015-12-01 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | FAK Forms a Complex with MEF2 to Couple Biomechanical Signaling to Transcription in Cardiomyocytes.
Structure, 24, 2016
|
|
3FEY
 
 | |
3FEX
 
 | |
5U0K
 
 | | C-terminal ankyrin repeats from human liver-type glutaminase (GAB/LGA) | | Descriptor: | Glutaminase liver isoform, mitochondrial | | Authors: | Ferreira, I.M, Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
5U0I
 
 | | C-terminal ankyrin repeats from human kidney-type glutaminase (KGA) - tetragonal crystal form | | Descriptor: | CHLORIDE ION, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
5UQE
 
 | | Multidomain structure of human kidney-type glutaminase(KGA/GLS) | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Pasquali, C.C, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2017-02-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
5U0J
 
 | | C-terminal ankyrin repeats from human kidney-type glutaminase (KGA) - monoclinic crystal form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, SODIUM ION | | Authors: | Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
8TNV
 
 | | Hemocyanin Functional Unit CCHB-g of Concholepas concholepas | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Munoz, S, Vallejos-Baccelliere, G, Manubens, A, Salazar, M, Nascimento, A.F.Z, Ambrosio, A.L.B, Becker, M.I, Guixe, V, Castro-Fernandez, V. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into a functional unit from an immunogenic mollusk hemocyanin.
Structure, 32, 2024
|
|
2B4J
 
 | | Structural basis for the recognition between HIV-1 integrase and LEDGF/p75 | | Descriptor: | GLYCEROL, Integrase (IN), PC4 and SFRS1 interacting protein, ... | | Authors: | Cherepanov, P, Ambrosio, A.L, Rahman, S, Ellenberger, T, Engelman, A. | | Deposit date: | 2005-09-24 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3SUL
 
 | | Crystal structure of cerato-platanin 3 from M. perniciosa (MpCP3) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUJ
 
 | | Crystal structure of cerato-platanin 1 from M. perniciosa (MpCP1) | | Descriptor: | ACETATE ION, CHLORIDE ION, Cerato-platanin 1, ... | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUK
 
 | | Crystal structure of cerato-platanin 2 from M. perniciosa (MpCP2) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3ST1
 
 | | Crystal structure of Necrosis and Ethylene inducing Protein 2 from the causal agent of cocoa's Witches Broom disease | | Descriptor: | Necrosis-and ethylene-inducing protein, SODIUM ION, ZINC ION | | Authors: | Oliveira, J.F, Zaparoli, G, Barsottini, M.R.O, Ambrosio, A.L.B, Pereira, G.A.G, Dias, S.M.G. | | Deposit date: | 2011-07-08 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Necrosis- and Ethylene-Inducing Protein 2 from the Causal Agent of Cacao's Witches' Broom Disease Reveals Key Elements for Its Activity.
Biochemistry, 50, 2011
|
|
