6P78
 
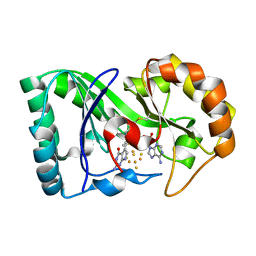 | | queuine lyase from Clostridium spiroforme bound to SAM and queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5S)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-1,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, IRON/SULFUR CLUSTER, Queuine lyase, ... | | Authors: | Almo, S.C, Grove, T.L. | | Deposit date: | 2019-06-05 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | Discovery of novel bacterial queuine salvage enzymes and pathways in human pathogens.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OVT
 
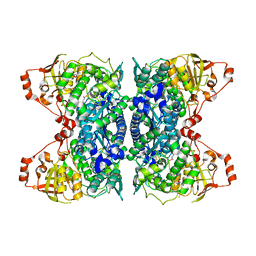 | | Crystal Structure of IlvD from Mycobacterium tuberculosis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dihydroxy-acid dehydratase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Almo, S.C, Grove, T.L, Bonanno, J.B, Baker, E.N, Bashiri, G. | | Deposit date: | 2019-05-08 | | Release date: | 2019-08-07 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The active site of theMycobacterium tuberculosisbranched-chain amino acid biosynthesis enzyme dihydroxyacid dehydratase contains a 2Fe-2S cluster.
J.Biol.Chem., 294, 2019
|
|
1AAW
 
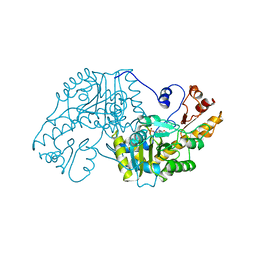 | | THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Almo, S.C, Smith, D.L, Danishefsky, A.T, Ringe, D. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli.
Protein Eng., 7, 1994
|
|
1AAM
 
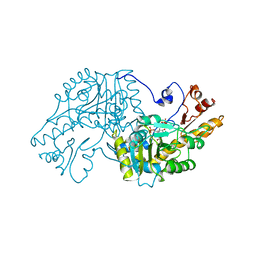 | | THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | Aspartate aminotransferase, SULFATE ION | | Authors: | Almo, S.C, Smith, D.L, Danishefsky, A.T, Ringe, D. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli.
Protein Eng., 7, 1994
|
|
7RBW
 
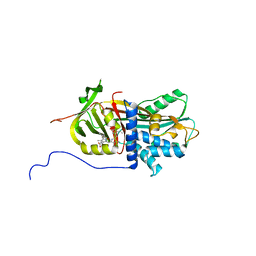 | | Structure of Biliverdin-binding Serpin of Boana punctata (polka-dot tree frog) | | Descriptor: | BILIVERDINE IX ALPHA, Biliverdin bindin serpin | | Authors: | Fedorov, E, Manoilov, K.Y, Verkhusha, V, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of a Biliverdin-Binding Near-Infrared Fluorescent Protein From the Serpin Superfamily.
J.Mol.Biol., 434, 2021
|
|
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
1I80
 
 | | CRYSTAL STRUCTURE OF M. TUBERCULOSIS PNP IN COMPLEX WITH IMINORIBITOL, 9-DEAZAHYPOXANTHINE AND PHOSPHATE ION | | Descriptor: | 9-DEAZAHYPOXANTHINE, IMINORIBITOL, PHOSPHATE ION, ... | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2001-03-12 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
1I85
 
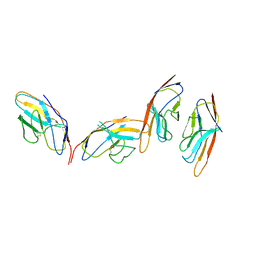 | | CRYSTAL STRUCTURE OF THE CTLA-4/B7-2 COMPLEX | | Descriptor: | CYTOTOXIC T-LYMPHOCYTE-ASSOCIATED PROTEIN 4, T LYMPHOCYTE ACTIVATION ANTIGEN CD86 | | Authors: | Schwartz, J.-C.D, Zhang, X, Fedorov, A.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2001-03-12 | | Release date: | 2001-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for co-stimulation by the human CTLA-4/B7-2 complex.
Nature, 410, 2001
|
|
8SFZ
 
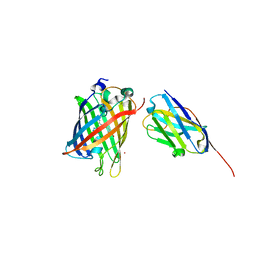 | | High Affinity nanobodies against GFP | | Descriptor: | Green fluorescent protein, LaG35, POTASSIUM ION, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unique mechanisms to increase structural stability and enhance antigen binding in nanobodies.
Structure, 33, 2025
|
|
8SFS
 
 | | High Affinity nanobodies against GFP | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonnano, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Unique mechanisms to increase structural stability and enhance antigen binding in nanobodies.
Structure, 33, 2025
|
|
6OFZ
 
 | | Crystal structure of human WDR5 | | Descriptor: | WD repeat-containing protein 5 | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OI1
 
 | | Crystal structure of human WDR5 in complex with monomethyl L-arginine | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OI2
 
 | | Crystal structure of human WDR5 in complex with symmetric dimethyl-L-arginine | | Descriptor: | GLYCEROL, N3, N4-DIMETHYLARGININE, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
7MSJ
 
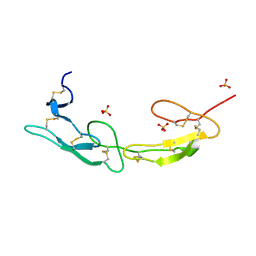 | | The crystal structure of mouse HVEM | | Descriptor: | SULFATE ION, Tumor necrosis factor receptor superfamily member 14 | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
7MSG
 
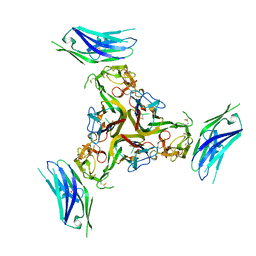 | | The crystal structure of LIGHT in complex with HVEM and CD160 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen, soluble form,Tumor necrosis factor receptor superfamily member 14, ... | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
6XI9
 
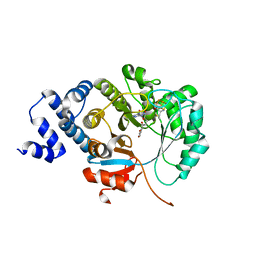 | | X-ray crystal structure of MqnE from Pedobacter heparinus in complex with aminofutalosine and methionine | | Descriptor: | 9-[7-(3-carboxyphenyl)-5,6-dideoxy-beta-D-ribo-heptodialdo-1,4-furanosyl]-9H-purin-6-amine, Aminodeoxyfutalosine synthase, CHLORIDE ION, ... | | Authors: | Grove, T.L, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Narrow-Spectrum Antibiotic Targeting of the Radical SAM Enzyme MqnE in Menaquinone Biosynthesis.
Biochemistry, 59, 2020
|
|
7MJV
 
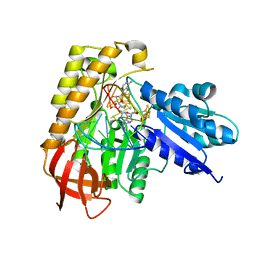 | | MiaB in the complex with s-adenosylmethionine and RNA | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJZ
 
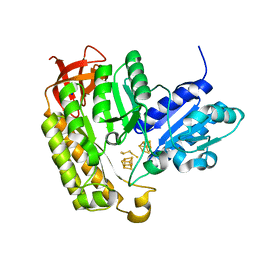 | | The structure of MiaB with pentasulfide bridge | | Descriptor: | IRON/SULFUR CLUSTER, PENTASULFIDE-SULFUR, SODIUM ION, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJY
 
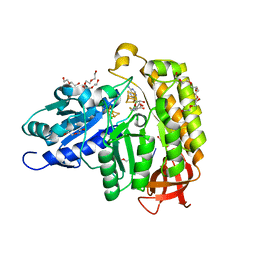 | | MiaB in the complex with s-adenosyl-L-homocysteine and RNA | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJW
 
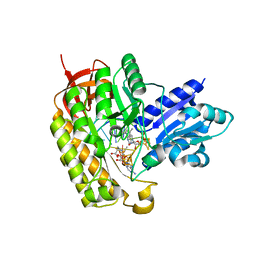 | | Methylated MiaB in the complex with 5'-deoxyadenosine, methionine and RNA | | Descriptor: | 5'-DEOXYADENOSINE, FE3-S4 methylated cluster, IRON/SULFUR CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJX
 
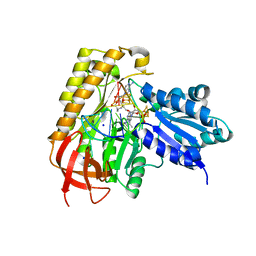 | | MiaB in the complex with 5'-deoxyadenosine, methionine and RNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-DEOXYADENOSINE, FE3-S4 CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
6NG3
 
 | | Crystal structure of human CD160 and HVEM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen,Tumor necrosis factor receptor superfamily member 14, MAGNESIUM ION, ... | | Authors: | Liu, W, Bonanno, J, Almo, S.C. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Basis of CD160:HVEM Recognition.
Structure, 27, 2019
|
|
6NG9
 
 | |
6NGG
 
 | |
1Z2I
 
 | |
