6OVT
 
 | | Crystal Structure of IlvD from Mycobacterium tuberculosis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dihydroxy-acid dehydratase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Almo, S.C, Grove, T.L, Bonanno, J.B, Baker, E.N, Bashiri, G. | | Deposit date: | 2019-05-08 | | Release date: | 2019-08-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The active site of theMycobacterium tuberculosisbranched-chain amino acid biosynthesis enzyme dihydroxyacid dehydratase contains a 2Fe-2S cluster.
J.Biol.Chem., 294, 2019
|
|
6P78
 
 | | queuine lyase from Clostridium spiroforme bound to SAM and queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5S)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-1,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, IRON/SULFUR CLUSTER, Queuine lyase, ... | | Authors: | Almo, S.C, Grove, T.L. | | Deposit date: | 2019-06-05 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | Discovery of novel bacterial queuine salvage enzymes and pathways in human pathogens.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1AAW
 
 | | THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Almo, S.C, Smith, D.L, Danishefsky, A.T, Ringe, D. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli.
Protein Eng., 7, 1994
|
|
1AAM
 
 | | THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Almo, S.C, Smith, D.L, Danishefsky, A.T, Ringe, D. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli.
Protein Eng., 7, 1994
|
|
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
1M9P
 
 | | Crystalline Human Carbonmonoxy Hemoglobin C Exhibits The R2 Quaternary State at Neutral pH In The Presence of Polyethylene Glycol: The 2.1 Angstrom Resolution Crystal Structure | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Patskovska, L.N, Patskovsky, Y.V, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2002-07-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | COHbC and COHbS crystallize in the R2 quaternary state at neutral pH in the presence of PEG 4000.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7UI0
 
 | | Post-fusion ectodomain of HSV-1 gB in complex with HSV010-13 Fab | | Descriptor: | Envelope glycoprotein B, HSV10-13 Fab Heavy chain, HSV10-13 Light chain | | Authors: | Windsor, I.W, Kong, S.L, Garforth, S.J, Almo, S.C, Harrison, S.C. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
J.Clin.Invest., 133, 2023
|
|
7UHZ
 
 | | Post-fusion ectodomain of HSV-1 gB in complex with BMPC-23 Fab | | Descriptor: | BMPC-23 Fab Heavy chain, BMPC-23 Fab Light chain, Envelope glycoprotein B | | Authors: | Windsor, I.W, Kong, S.L, Garforth, S.J, Almo, S.C, Harrison, S.C. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
J.Clin.Invest., 133, 2023
|
|
7RBW
 
 | | Structure of Biliverdin-binding Serpin of Boana punctata (polka-dot tree frog) | | Descriptor: | BILIVERDINE IX ALPHA, Biliverdin bindin serpin | | Authors: | Fedorov, E, Manoilov, K.Y, Verkhusha, V, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of a Biliverdin-Binding Near-Infrared Fluorescent Protein From the Serpin Superfamily.
J.Mol.Biol., 434, 2021
|
|
6NG3
 
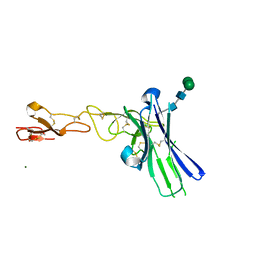 | | Crystal structure of human CD160 and HVEM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen,Tumor necrosis factor receptor superfamily member 14, MAGNESIUM ION, ... | | Authors: | Liu, W, Bonanno, J, Almo, S.C. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Basis of CD160:HVEM Recognition.
Structure, 27, 2019
|
|
6NGG
 
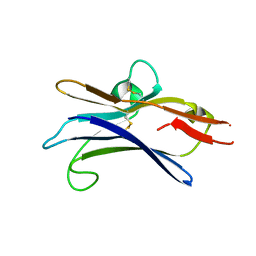 | |
6NG9
 
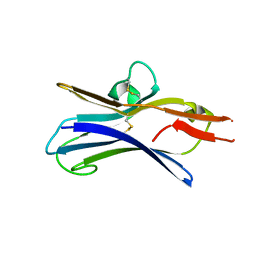 | |
5VG6
 
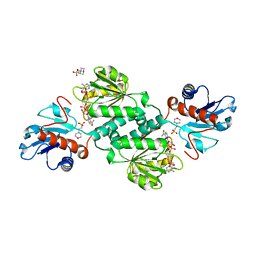 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 in complex with NADPH and MES. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lipowska, J, Shabalin, I.G, Kutner, J, Gasiorowska, O.A, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-04-10 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 in complex with NADPH and MES.
to be published
|
|
6XIG
 
 | |
6XI9
 
 | | X-ray crystal structure of MqnE from Pedobacter heparinus in complex with aminofutalosine and methionine | | Descriptor: | 9-[7-(3-carboxyphenyl)-5,6-dideoxy-beta-D-ribo-heptodialdo-1,4-furanosyl]-9H-purin-6-amine, Aminodeoxyfutalosine synthase, CHLORIDE ION, ... | | Authors: | Grove, T.L, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Narrow-Spectrum Antibiotic Targeting of the Radical SAM Enzyme MqnE in Menaquinone Biosynthesis.
Biochemistry, 59, 2020
|
|
4UC0
 
 | | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Sampathkumar, P, Ramagopal, U.A, Attonito, J, Ahmed, M, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
To be published
|
|
4TYM
 
 | | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935 | | Descriptor: | Purine nucleoside phosphorylase DeoD-type, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935.
To Be Published
|
|
4U13
 
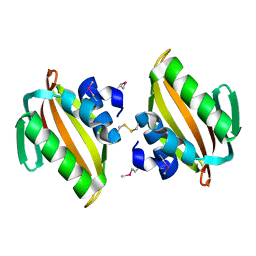 | | Crystal structure of putative polyketide cyclase (protein SMa1630) from Sinorhizobium meliloti at 2.3 A resolution | | Descriptor: | putative polyketide cyclase SMa1630 | | Authors: | Shabalin, I.G, Bacal, P, Osinski, T, Cooper, D.R, Szlachta, K, Stead, M, Grabowski, M, Hammonds, J, Ahmed, M, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-07-14 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative polyketide cyclase (protein SMa1630) from Sinorhizobium meliloti at 2.3 A resolution
to be published
|
|
4UAB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound ethanolamine | | Descriptor: | CHLORIDE ION, ETHANOLAMINE, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-08 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
5KZK
 
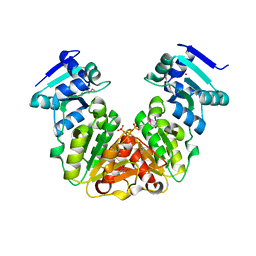 | | Crystal Structure of rRNA methyltransferase from Sinorhizobium meliloti | | Descriptor: | COBALT (II) ION, Probable RNA methyltransferase, TrmH family, ... | | Authors: | Dey, D, Hegde, R.P, Almo, S.C, Ramakumar, S, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-07-25 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of rRNA methyltransferase from Sinorhizobium meliloti
To Be Published
|
|
5L19
 
 | | Crystal Structure of a human FasL mutant | | Descriptor: | SULFATE ION, Tumor necrosis factor ligand superfamily member 6, ZINC ION | | Authors: | Liu, W, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2016-07-28 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3.
Structure, 24, 2016
|
|
5L0Z
 
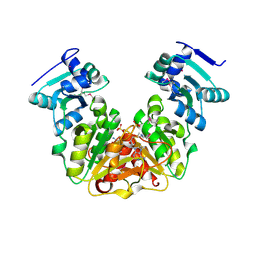 | | Crystal Structure of AdoMet bound rRNA methyltransferase from Sinorhizobium meliloti | | Descriptor: | COBALT (II) ION, Probable RNA methyltransferase, TrmH family, ... | | Authors: | Dey, D, Hegde, R.P, Almo, S.C, Ramakumar, S, Ramagopal, U.A. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of AdoMet bound rRNA methyltransferase from Sinorhizobium meliloti
To Be Published
|
|
5L36
 
 | |
7N7I
 
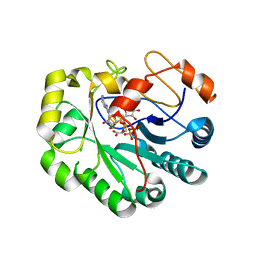 | | X-ray crystal structure of Viperin-like enzyme from Trichoderma virens | | Descriptor: | IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Grove, T.L, Almo, S.C, Bonanno, J.B, Lachowicz, J.C, Gizzi, A.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural Insight into the Substrate Scope of Viperin and Viperin-like Enzymes from Three Domains of Life.
Biochemistry, 60, 2021
|
|
7N2W
 
 | | The crystal structure of an FMN-dependent NADH-azoreductase, AzoA in complex with Red 40 | | Descriptor: | 6-hydroxy-5-[(E)-(2-methoxy-5-methyl-4-sulfophenyl)diazenyl]naphthalene-2-sulfonic acid, FLAVIN MONONUCLEOTIDE, FMN dependent NADH:quinone oxidoreductase | | Authors: | Arcinas, A.J, Fedorov, E, Kelly, L, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Uncovering a novel mechanism of enzyme activation in multimeric azoreductases
To Be Published
|
|
