8WJY
 
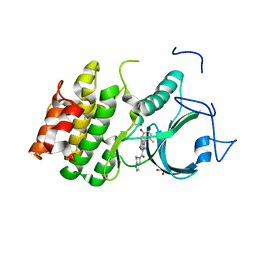 | | PKMYT1_Cocrystal_Cpd 4 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Wang, Y, Wang, C, Liu, T, Qi, H, Chen, S, Cai, X, Zhang, M, Aliper, A, Ren, F, Ding, X, Zhavoronkov, A. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of Tetrahydropyrazolopyrazine Derivatives as Potent and Selective MYT1 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
6D35
 
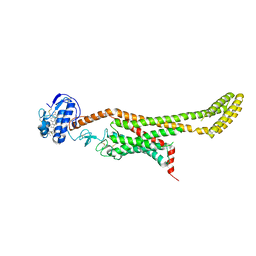 | | Crystal structure of Xenopus Smoothened in complex with cholesterol | | Descriptor: | CHOLESTEROL, Smoothened,Soluble cytochrome b562,Smoothened | | Authors: | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | Deposit date: | 2018-04-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
6D32
 
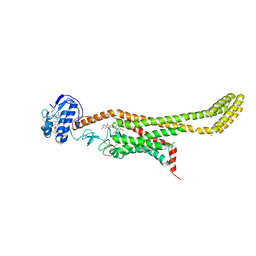 | | Crystal structure of Xenopus Smoothened in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened,Soluble cytochrome b562,Smoothened | | Authors: | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | Deposit date: | 2018-04-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
2BK5
 
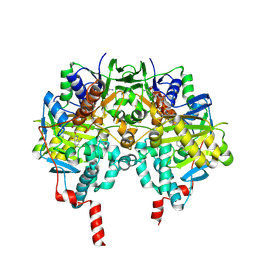 | | Human Monoamine Oxidase B: I199F mutant in complex with isatin | | Descriptor: | AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE, ISATIN | | Authors: | Binda, C, Edmondson, D.E, Mattevi, A, Hubalek, F, Khalil, A, Li, M, Castagnoli, N. | | Deposit date: | 2005-02-10 | | Release date: | 2005-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Demonstration of Isoleucine 199 as a Structural Determinant for the Selective Inhibition of Human Monoamine Oxidase B by Specific Reversible Inhibitors.
J.Biol.Chem., 280, 2005
|
|
7ZVY
 
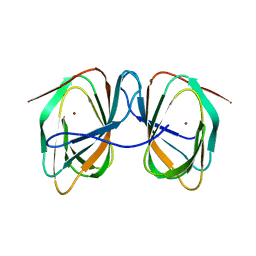 | | Thermococcus kadokarensis phosphomannose isomerase | | Descriptor: | Cupin_2 domain-containing protein, ZINC ION | | Authors: | Hoh, f, Calio, A. | | Deposit date: | 2022-05-17 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Unravelling the Adaptation Mechanisms to High Pressure in Proteins.
Int J Mol Sci, 23, 2022
|
|
7ZVM
 
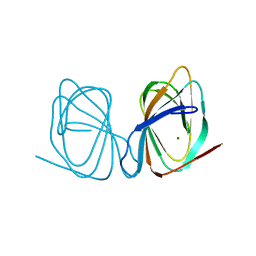 | |
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
2BK4
 
 | | Human Monoamine Oxidase B: I199F mutant in complex with rasagiline | | Descriptor: | (1R)-N-(prop-2-en-1-yl)-2,3-dihydro-1H-inden-1-amine, AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Edmondson, D.E, Mattevi, A, Hubalek, F, Khalil, A, Li, M, Castagnoli, N. | | Deposit date: | 2005-02-10 | | Release date: | 2005-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Demonstration of Isoleucine 199 as a Structural Determinant for the Selective Inhibition of Human Monoamine Oxidase B by Specific Reversible Inhibitors
J.Biol.Chem., 280, 2005
|
|
6DRD
 
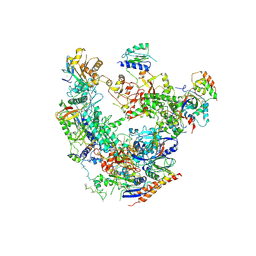 | | RNA Pol II(G) | | Descriptor: | DNA-directed RNA polymerase II subunit GRINL1A, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Yu, X, Jishage, M, Shi, Y, Ganesan, S, Sali, A, Chait, B.T, Asturias, F, Roeder, R.G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-12 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3DFA
 
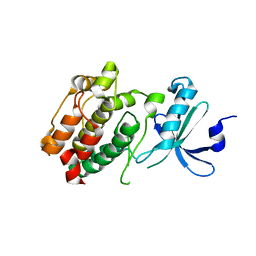 | | Crystal structure of kinase domain of calcium-dependent protein kinase cgd3_920 from Cryptosporidium parvum | | Descriptor: | Calcium-dependent protein kinase cgd3_920 | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Hassanali, A, Khuu, C, Alam, Z, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wilkstrom, M, Edwards, A.M, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-11 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of kinase domain of calcium-dependent protein kinase cgd3_920 from Cryptosporidium parvum.
To be Published
|
|
1F2P
 
 | | CRYSTAL STRUCTURE OF THE STREPTOMYCES GRISEUS AMINOPEPTIDASE COMPLEXED WITH L-PHENYLALANINE | | Descriptor: | AMINOPEPTIDASE, CALCIUM ION, PHENYLALANINE, ... | | Authors: | Gilboa, R, Spungin-Bialik, A, Wohlfahrt, G, Schomburg, D, Blumberg, S, Shoham, G. | | Deposit date: | 2000-05-28 | | Release date: | 2001-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions of Streptomyces griseus aminopeptidase with amino acid reaction products and their implications toward a catalytic mechanism.
Proteins, 44, 2001
|
|
1F2O
 
 | | CRYSTAL STRUCTURE OF THE STREPTOMYCES GRISEUS AMINOPEPTIDASE COMPLEXED WITH L-LEUCINE | | Descriptor: | AMINOPEPTIDASE, CALCIUM ION, LEUCINE, ... | | Authors: | Gilboa, R, Spungin-Bialik, A, Wohlfahrt, G, Schomburg, D, Blumberg, S, Shoham, G. | | Deposit date: | 2000-05-28 | | Release date: | 2001-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of Streptomyces griseus aminopeptidase with amino acid reaction products and their implications toward a catalytic mechanism.
Proteins, 44, 2001
|
|
3C3E
 
 | | Crystal structure of 2-phospho-(S)-lactate transferase from Methanosarcina mazei in complex with Fo and GDP. Northeast Structural Genomics Consortium target MaR46 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, 2-phospho-L-lactate transferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Forouhar, F, Abashidze, M, Xu, H, Grochowski, L.L, Seetharaman, J, Hussain, M, Kuzin, A.P, Chen, Y, Zhou, W, Xiao, R, Acton, T.B, Montelione, G.T, Galinier, A, White, R.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular insights into the biosynthesis of the f420 coenzyme.
J.Biol.Chem., 283, 2008
|
|
7P7P
 
 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide((1R)-1-Amino-3-phenylpropyl){(2S)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)amino]-2-{[3-(2-hydroxyphenyl)-isoxazol-5-yl]methyl}-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
7PFS
 
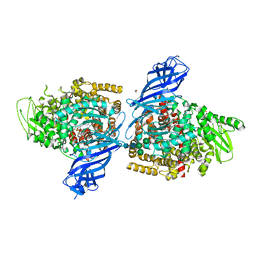 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide ((1R)-1-Amino-3-phenylpropyl){2-([1,1:3,1-terphenyl]-5-ylmethyl)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)-amino]-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
1JB9
 
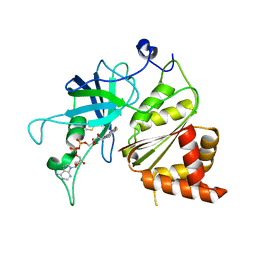 | | Crystal Structure of The Ferredoxin:NADP+ Reductase From Maize Root AT 1.7 Angstroms | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, ferredoxin-NADP reductase | | Authors: | Faber, H.R, Karplus, P.A, Aliverti, A, Ferioli, C, Spinola, M. | | Deposit date: | 2001-06-03 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues.
Biochemistry, 40, 2001
|
|
1K1C
 
 | | Solution Structure of Crh, the Bacillus subtilis Catabolite Repression HPr | | Descriptor: | catabolite repression HPr-like protein | | Authors: | Favier, A, Brutscher, B, Blackledge, M, Galinier, A, Deutscher, J, Penin, F, Marion, D. | | Deposit date: | 2001-09-25 | | Release date: | 2001-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Crh, the Bacillus subtilis catabolite repression HPr.
J.Mol.Biol., 317, 2002
|
|
1LQT
 
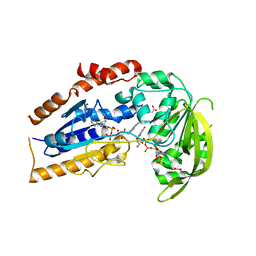 | | A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase | | Descriptor: | 4-OXO-NICOTINAMIDE-ADENINE DINUCLEOTIDE PHOSPHATE, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bossi, R.T, Aliverti, A, Raimondi, D, Fischer, F, Zanetti, G, Ferrari, D, Tahallah, N, Maier, C.S, Heck, A.J.R, Rizzi, M, Mattevi, A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-13 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase.
Biochemistry, 41, 2002
|
|
1LQU
 
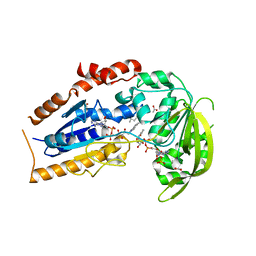 | | Mycobacterium tuberculosis FprA in complex with NADPH | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, FprA, ... | | Authors: | Bossi, R.T, Aliverti, A, Raimondi, D, Fischer, F, Zanetti, G, Ferrari, D, Tahallah, N, Maier, C.S, Heck, A.J.R, Rizzi, M, Mattevi, A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-13 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase.
Biochemistry, 41, 2002
|
|
1LR1
 
 | | Solution Structure of the Oligomerization Domain of the Bacterial Chromatin-Structuring Protein H-NS | | Descriptor: | dna-binding protein h-ns | | Authors: | Esposito, D, Petrovic, A, Harris, R, Ono, S, Eccleston, J, Mbabaali, A, Haq, I, Higgins, C.F, Hinton, J.C.D, Driscoll, P.C, Ladbury, J.E. | | Deposit date: | 2002-05-14 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | H-NS Oligomerization Domain Structure Reveals the Mechanism for High Order
Self-association of the Intact Protein
J.Mol.Biol., 324, 2002
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
5FM1
 
 | | Structure of gamma-tubulin small complex based on a cryo-EM map, chemical cross-links, and a remotely related structure | | Descriptor: | SPINDLE POLE BODY COMPONENT 110, SPINDLE POLE BODY COMPONENT SPC97, SPINDLE POLE BODY COMPONENT SPC98, ... | | Authors: | Greenberg, C.H, Kollman, J, Zelter, A, Johnson, R, MacCoss, M.J, Davis, T.N, Agard, D.A, Sali, A. | | Deposit date: | 2015-10-30 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure of Gamma-Tubulin Small Complex Based on a Cryo-Em Map, Chemical Cross-Links, and a Remotely Related Structure.
J.Struct.Biol., 194, 2016
|
|
5FLZ
 
 | | Cryo-EM structure of gamma-TuSC oligomers in a closed conformation | | Descriptor: | SPINDLE POLE BODY COMPONENT 110, SPINDLE POLE BODY COMPONENT SPC97, SPINDLE POLE BODY COMPONENT SPC98, ... | | Authors: | Greenberg, C.H, Kollman, J, Zelter, A, Johnson, R, MacCoss, M.J, Davis, T.N, Agard, D.A, Sali, A. | | Deposit date: | 2015-10-29 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of Gamma-Tubulin Small Complex Based on a Cryo-Em Map, Chemical Cross-Links, and a Remotely Related Structure.
J.Struct.Biol., 194, 2016
|
|
