2FEC
 
 | | Structure of the E203Q mutant of the Cl-/H+ exchanger CLC-ec1 from E.Coli | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Accardi, A, Walden, M.P, Nguitragool, W, Jayaram, H, Williams, C, Miller, C. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.967 Å) | | Cite: | Separate ion pathways in a Cl-/H+ exchanger
J.Gen.Physiol., 126, 2005
|
|
2FEE
 
 | | Structure of the Cl-/H+ exchanger CLC-ec1 from E.Coli in NaBr | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Accardi, A, Walden, M.P, Nguitragool, W, Jayaram, H, Williams, C, Miller, C. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Separate ion pathways in a Cl-/H+ exchanger
J.Gen.Physiol., 126, 2005
|
|
2FED
 
 | | Structure of the E203Q mutant of the Cl-/H+ exchanger CLC-ec1 from E.Coli | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Accardi, A, Walden, M.P, Nguitragool, W, Jayaram, H, Williams, C, Miller, C. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.317 Å) | | Cite: | Separate ion pathways in a Cl-/H+ exchanger
J.Gen.Physiol., 126, 2005
|
|
2HT2
 
 | | Structure of the Escherichia coli ClC chloride channel Y445H mutant and Fab complex | | Descriptor: | BROMIDE ION, Fab fragment, heavy chain, ... | | Authors: | Accardi, A, Lobet, S, Williams, C, Miller, C, Dutzler, R. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Synergism Between Halide Binding and Proton Transport in a CLC-type Exchanger.
J.Mol.Biol., 362, 2006
|
|
2HTL
 
 | | Structure of the Escherichia coli ClC chloride channel Y445F mutant and Fab complex | | Descriptor: | BROMIDE ION, Fab fragment, Heavy chain, ... | | Authors: | Accardi, A, Lobet, S, Williams, C, Miller, C, Dutzler, R. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Synergism Between Halide Binding and Proton Transport in a CLC-type Exchanger.
J.Mol.Biol., 362, 2006
|
|
2HLF
 
 | | Structure of the Escherichis coli ClC chloride channel Y445E mutant and Fab complex | | Descriptor: | BROMIDE ION, Fab Fragment, Heavy chain, ... | | Authors: | Accardi, A, Lobet, S, Williams, C, Miller, C, Dutzler, R. | | Deposit date: | 2006-07-07 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Synergism Between Halide Binding and Proton Transport in a CLC-type Exchanger
J.Mol.Biol., 362, 2006
|
|
2HT3
 
 | | Structure of the Escherichia coli ClC chloride channel Y445L mutant and Fab complex | | Descriptor: | BROMIDE ION, Fab fragment, Heavy chain, ... | | Authors: | Accardi, A, Lobet, S, Williams, C, Miller, C, Dutzler, R. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Synergism between halide binding and proton transport in a CLC-type exchanger
J.Mol.Biol., 362, 2006
|
|
2HT4
 
 | | Structure of the Escherichia coli ClC chloride channel Y445W mutant and Fab complex | | Descriptor: | BROMIDE ION, Fab fragment, Heavy chain, ... | | Authors: | Accardi, A, Lobet, S, Williams, C, Miller, C, Dutzler, R. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Synergism Between Halide Binding and Proton Transport in a CLC-type Exchanger.
J.Mol.Biol., 362, 2006
|
|
2HTK
 
 | | Structure of the Escherichia coli ClC chloride channel Y445A mutant and Fab complex | | Descriptor: | BROMIDE ION, Fab fragment, Heavy chain, ... | | Authors: | Accardi, A, Lobet, S, Williams, C, Miller, C, Dutzler, R. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Synergism between halide binding and proton transport in a CLC-type exchanger
J.Mol.Biol., 362, 2006
|
|
6E1O
 
 | |
6DZ7
 
 | |
6E0H
 
 | |
8TPO
 
 | |
6OY3
 
 | | nhTMEM16 L302A +Ca2+ in nanodiscs | | Descriptor: | CALCIUM ION, nhTMEM16 | | Authors: | Falzone, M, Lee, B.C, Accardi, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Dynamic modulation of the lipid translocation groove generates a conductive ion channel in Ca 2+ -bound nhTMEM16.
Nat Commun, 10, 2019
|
|
3DET
 
 | | Structure of the E148A, Y445A doubly ungated mutant of E.coli CLC_Ec1, Cl-/H+ antiporter | | Descriptor: | Fab fragment, Heavy chain, Light chain, ... | | Authors: | Jayaram, H, Accardi, A, Wu, F, Williams, C, Miller, C. | | Deposit date: | 2008-06-10 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ion permeation through a Cl--selective channel designed from a CLC Cl-/H+ exchanger
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7RXH
 
 | |
7RXG
 
 | |
7RX2
 
 | |
7RXA
 
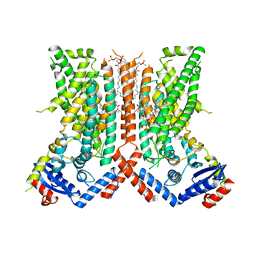 | | afTMEM16 DE/AA mutant in C14 lipid nanodiscs in the presence of Ca2+ | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, afTMEM16 lipid scramblase | | Authors: | Feng, Z, Accardi, A. | | Deposit date: | 2021-08-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | TMEM16 scramblases thin the membrane to enable lipid scrambling.
Nat Commun, 13, 2022
|
|
7RXB
 
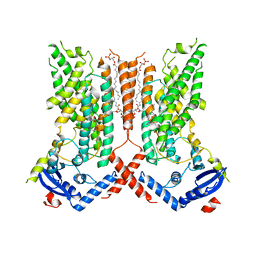 | | afTMEM16 lipid scramblase in C18 lipid nanodiscs in the absence of Ca2+ | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, afTMEM16 lipid scramblase | | Authors: | Feng, Z, Accardi, A. | | Deposit date: | 2021-08-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | TMEM16 scramblases thin the membrane to enable lipid scrambling.
Nat Commun, 13, 2022
|
|
7RX3
 
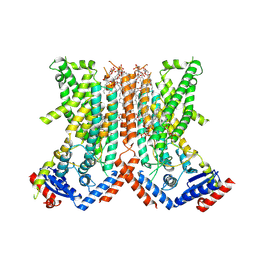 | |
7RWJ
 
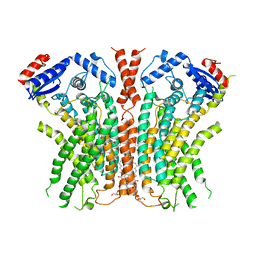 | |
7N8P
 
 | | CLC-ec1 at pH 4.5 100mM Cl Swap | | Descriptor: | H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Boudker, O, Accardi, A. | | Deposit date: | 2021-06-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural basis of common gate activation in CLC transporters
To Be Published
|
|
7N9W
 
 | | CLC-ec1 pH 4.5 100mM Cl Twist1 | | Descriptor: | H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Boudker, O, Accardi, A. | | Deposit date: | 2021-06-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural basis of common gate activation in CLC transporters
To Be Published
|
|
8GA0
 
 | | CLC-ec1 E202Y at pH 4.5 100mM Cl Turn | | Descriptor: | H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Lee, S, Argyos, Y, Chadda, R, Ciftci, D, Huysmans, G, Robertson, J.L, Boudker, O, Accardi, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of pH-dependent activation in a CLC transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
