4N8W
 
 | | cathepsin K - chondroitin sulfate complex | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, Cathepsin K | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2013-10-18 | | Release date: | 2014-11-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis of collagen fiber degradation by cathepsin K.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MT2
 
 | |
4Q30
 
 | | Nitrowillardiine bound to the ligand binding domain of GluA2 at pH 3.5 | | Descriptor: | 3-(5-nitro-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-L-alanine, Glutamate receptor 2 CHIMERIC PROTEIN, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2014-04-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Thermodynamics and mechanism of the interaction of willardiine partial agonists with a glutamate receptor: implications for drug development.
Biochemistry, 53, 2014
|
|
4QIG
 
 | |
4QIV
 
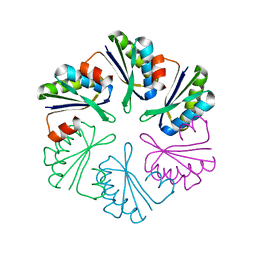 | |
4KPC
 
 | | Crystal structure of the nucleoside diphosphate kinase b from Leishmania braziliensis | | Descriptor: | Nucleoside diphosphate kinase b, PHOSPHATE ION | | Authors: | Vieira, P.S, Giuseppe, P.O, Santos, C.R, Cunha, E.M.F, de Oliveira, A.H.C, Murakami, M.T. | | Deposit date: | 2013-05-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and biophysical characterization of the nucleoside diphosphate kinase from Leishmania braziliensis.
Bmc Struct.Biol., 15, 2015
|
|
4KT8
 
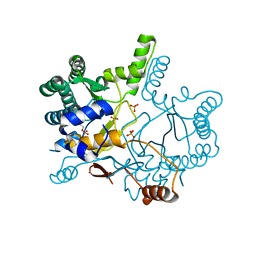 | | The complex structure of Rv3378c-Y51FY90F with substrate, TPP | | Descriptor: | (2E)-3-methyl-5-[(1R,2S,8aS)-1,2,5,5-tetramethyl-1,2,3,5,6,7,8,8a-octahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
4QI1
 
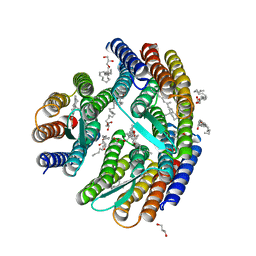 | | Crystal structure of H. walsbyi bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin-I, GLYCEROL, RETINAL, ... | | Authors: | Wang, A.H.J, Hsu, M.F, Yang, C.S, Fu, H.Y. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Studies of a Newly Grouped Haloquadratum walsbyi Bacteriorhodopsin Reveal the Acid-resistant Light-driven Proton Pumping Activity.
J. Biol. Chem., 290, 2015
|
|
4QID
 
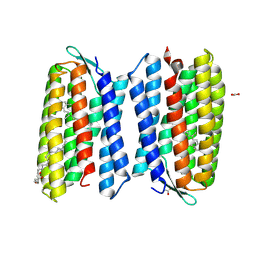 | | Crystal structure of Haloquadratum walsbyi bacteriorhodopsin | | Descriptor: | ACETATE ION, Bacteriorhodopsin-I, RETINAL, ... | | Authors: | Wang, A.H.J, Hsu, M.F, Yang, C.S, Fu, H.Y. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | An acid-tolerant light-driven proton pump
To be Published
|
|
4K7J
 
 | | Peptidoglycan O-acetylesterase in action, 5 min | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, GDSL-like Lipase/Acylhydrolase family protein | | Authors: | Williams, A.H, Gomperts Boneca, I. | | Deposit date: | 2013-04-17 | | Release date: | 2014-09-03 | | Last modified: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Visualization of a substrate-induced productive conformation of the catalytic triad of the Neisseria meningitidis peptidoglycan O-acetylesterase reveals mechanistic conservation in SGNH esterase family members.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4K3U
 
 | | Peptidoglycan O-acetylesterase in action, 30 min | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GDSL-like Lipase/Acylhydrolase family protein | | Authors: | Williams, A.H, Gomperts Boneca, I. | | Deposit date: | 2013-04-11 | | Release date: | 2014-09-03 | | Last modified: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (2.158 Å) | | Cite: | Visualization of a substrate-induced productive conformation of the catalytic triad of the Neisseria meningitidis peptidoglycan O-acetylesterase reveals mechanistic conservation in SGNH esterase family members.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QIE
 
 | |
4MHZ
 
 | | Crystal structure of apo-form glutaminyl cyclase from Ixodes scapularis in complex with PBD150 | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, Glutaminyl cyclase, putative | | Authors: | Huang, K.F, Hsu, H.L, Wang, A.H.J. | | Deposit date: | 2013-08-30 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional analyses of a glutaminyl cyclase from Ixodes scapularis reveal metal-independent catalysis and inhibitor binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QNN
 
 | |
3H03
 
 | |
3H5Z
 
 | | Crystal Structure of Leishmania major N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, ... | | Authors: | Qiu, W, Hutchinson, A, Lin, Y.-H, Wernimont, A, Mackenzie, F, Ravichandran, M, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Fairlamb, A.H, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | N-myristoyltransferase inhibitors as new leads to treat sleeping sickness.
Nature, 464, 2010
|
|
3HS4
 
 | | Human carbonic anhydrase II complexed with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Robbins, A.H, Genis, C, Domsic, J, Sippel, K.H, Agbandje-McKenna, M, McKenna, R. | | Deposit date: | 2009-06-10 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution structure of human carbonic anhydrase II complexed with acetazolamide reveals insights into inhibitor drug design.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3FWN
 
 | | Dimeric 6-phosphogluconate dehydrogenase complexed with 6-phosphogluconate and 2'-monophosphoadenosine-5'-diphosphate | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconate dehydrogenase, ... | | Authors: | Chen, Y.-Y, Ko, T.-P, Lo, L.-P, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2009-01-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism
J.Struct.Biol., 169, 2010
|
|
3HZD
 
 | | Crystal structure of bothropstoxin-I (BthTX-I), a PLA2 homologue from Bothrops jararacussu venom | | Descriptor: | LITHIUM ION, Phospholipase A2 homolog bothropstoxin-1 | | Authors: | Silva, M.C.O, Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
3I03
 
 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) - monomeric form at a high resolution | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
3HTL
 
 | | Structure of the Corynebacterium diphtheriae major pilin SpaA points to a modular pilus assembly with stabilizing isopeptide bonds | | Descriptor: | CALCIUM ION, Putative surface-anchored fimbrial subunit, SODIUM ION | | Authors: | Kang, H.J, Paterson, N.G, Gaspar, A.H, Ton-That, H, Baker, E.N. | | Deposit date: | 2009-06-11 | | Release date: | 2009-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3I3A
 
 | | Structural Basis for the Sugar Nucleotide and Acyl Chain Selectivity of Leptospira interrogans LpxA | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, S-[2-({N-[(2R)-2-hydroxy-4-{[(S)-hydroxy(methoxy)phosphoryl]oxy}-3,3-dimethylbutanoyl]-beta-alanyl}amino)ethyl] (3R)-3-hydroxydodecanethioate | | Authors: | Williams, A.H. | | Deposit date: | 2009-06-30 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for the sugar nucleotide and acyl-chain selectivity of Leptospira interrogans LpxA.
Biochemistry, 48, 2009
|
|
3HR6
 
 | | Structure of the Corynebacterium diphtheriae major pilin SpaA points to a modular pilus assembly stabilizing isopeptide bonds | | Descriptor: | CALCIUM ION, IODIDE ION, Putative surface-anchored fimbrial subunit | | Authors: | Kang, H.J, Paterson, N.G, Gaspar, A.H, Ton-That, H, Baker, E.N. | | Deposit date: | 2009-06-08 | | Release date: | 2009-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3I3X
 
 | |
3HSQ
 
 | |
