4U2G
 
 | | Crystal structure of dienelactone hydrolase B-4 variant (Q35H, F38L, Y64H, Q76L, Q110L, C123S, Y137C, A141V, Y145C, N154D, E199G, S208G, G211D, S233G and 237Q) at 1.80 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4YM9
 
 | | Crystal structure of Porcine Pancreatic Elastase (PPE) in complex with the novel inhibitor JM102 | | Descriptor: | 2-ethyl-2-(hydroxymethyl)-N-(6-methylpyridin-3-yl)butanamide, ACETATE ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Hofbauer, S, Brito, J.A, Mulchande, J, Nogly, P, Pessanha, M, Moreira, R, Archer, M. | | Deposit date: | 2015-03-06 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Clickable 4-Oxo-beta-lactam-Based Selective Probing for Human Neutrophil Elastase Related Proteomes.
ChemMedChem, 11, 2016
|
|
5O02
 
 | |
6SRP
 
 | |
7LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
6MLD
 
 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) F52A mutant from Salmonella typhimurium | | Descriptor: | ACETATE ION, Lysine/arginine/ornithine-binding periplasmic protein | | Authors: | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2018-09-27 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
3N94
 
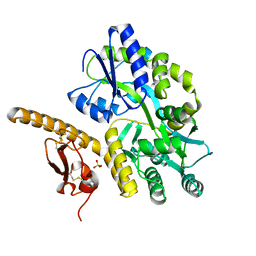 | | Crystal structure of human pituitary adenylate cyclase 1 Receptor-short N-terminal extracellular domain | | Descriptor: | Fusion protein of Maltose-binding periplasmic protein and pituitary adenylate cyclase 1 Receptor-short, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kumar, S, Pioszak, A.A, Swaminathan, K, Xu, H.E. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the PAC1R Extracellular Domain Unifies a Consensus Fold for Hormone Recognition by Class B G-Protein Coupled Receptors.
Plos One, 6, 2011
|
|
5HGI
 
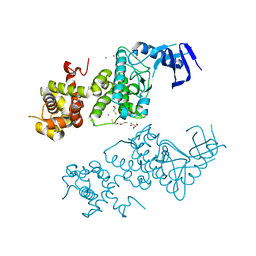 | | Crystal structure of apo human IRE1 alpha | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-MERCAPTOETHANOL, CESIUM ION, ... | | Authors: | Feldman, H.C, Tong, M, Wang, L, Meza-Acevedo, R, Gobillot, T.A, Gliedt, J.M, Hari, S.B, Mitra, A.K, Backes, B.J, Papa, F.R, Seeliger, M.A, Maly, D.J. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.584 Å) | | Cite: | Structural and Functional Analysis of the Allosteric Inhibition of IRE1 alpha with ATP-Competitive Ligands.
Acs Chem.Biol., 11, 2016
|
|
6FTA
 
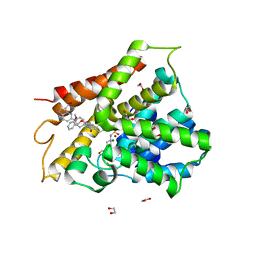 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-3098 | | Descriptor: | (4~{a}~{S},8~{a}~{S})-4-[4-methoxy-3-[(2-methoxyphenyl)methoxy]phenyl]-2-[1-(3-nitroimidazo[1,2-b]pyridazin-6-yl)piperidin-4-yl]-4~{a},5,6,7,8,8~{a}-hexahydrophthalazin-1-one, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-02-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-3098
To be published
|
|
5IU1
 
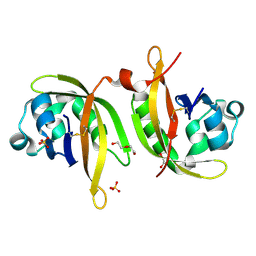 | | N-terminal PAS domain homodimer of PpANR MAP3K from Physcomitrella patens. | | Descriptor: | CTR1-like protein, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Stevenson, R.R, Trinh, C.H, Edwards, T.A, Cuming, A.C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Genetic Analysis of Physcomitrella patens Identifies ABSCISIC ACID NON-RESPONSIVE, a Regulator of ABA Responses Unique to Basal Land Plants and Required for Desiccation Tolerance.
Plant Cell, 28, 2016
|
|
8SJK
 
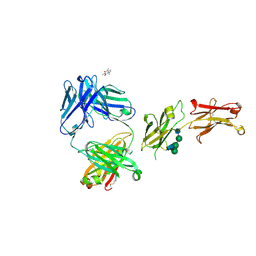 | | Pembrolizumab Caffeine crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY HEAVY CHAIN, ANTIBODY LIGHT CHAIN, ... | | Authors: | Larpent, P, Codan, L, Bothe, J.R, Stueber, D, Reichert, P, Fischmann, T, Su, Y, Pabit, S, Gupta, S, Iuzzolino, L, Cote, A. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Small-Angle X-ray Scattering as a Powerful Tool for Phase and Crystallinity Assessment of Monoclonal Antibody Crystallites in Support of Batch Crystallization.
Mol Pharm., 21, 2024
|
|
5O3P
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803 in Apo state, trigonal crystal form | | Descriptor: | DI(HYDROXYETHYL)ETHER, Membrane-associated protein slr1513, PENTAETHYLENE GLYCOL | | Authors: | Selim, K.A, Albrecht, R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2017-05-24 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PII-like signaling protein SbtB links cAMP sensing with cyanobacterial inorganic carbon response.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7TBH
 
 | | cryo-EM structure of MBP-KIX-apoferritin complex with peptide 7 | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed, LEU-SER-ARG-ARG-PRO-SEP-TYR-ARG-LYS-ILE-LEU-ASN-ASP-LEU-SER-SER-ASP-ALA-PRO | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
6SRL
 
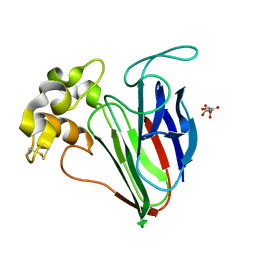 | |
8YPK
 
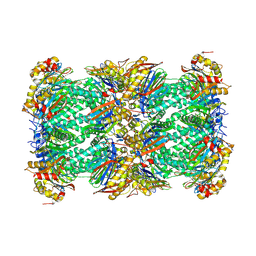 | | mouse proteasome 20S subunit in complex with compound 1 | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Kashima, A, Arai, Y. | | Deposit date: | 2024-03-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Optimization of alpha-amido boronic acids via cryo-electron microscopy analysis: Discovery of a novel highly selective immunoproteasome subunit LMP7 ( beta 5i)/LMP2 ( beta 1i) dual inhibitor.
Bioorg.Med.Chem., 109, 2024
|
|
7UFX
 
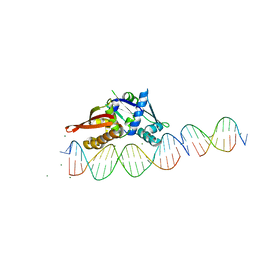 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and scaffold-insert duplexes (21mer and 10mer) containing the cognate REPE54 sequence and an additional G-C rich sequence, respectively. | | Descriptor: | DNA (5'-D(A*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*A)-3'), DNA (5'-D(A*GP*AP*CP*GP*GP*CP*CP*CP*GP*G)-3'), DNA (5'-D(A*GP*TP*CP*CP*GP*GP*GP*CP*CP*G)-3'), ... | | Authors: | Ward, A.R, Shields, E.T, Snow, C.D. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
8QG0
 
 | | Archaeoglobus fulgidus AfAgo complex with AfAgo-N protein (fAfAgo) bound with 17 nt RNA guide and 17 nt DNA target | | Descriptor: | AfAgo-N protein, DNA target 17 nt, Piwi protein, ... | | Authors: | Manakova, E.N, Zaremba, M, Pocevicuite, R, Golovinas, E, Zagorskaite, E, Silanskas, A. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res., 52, 2024
|
|
3N7L
 
 | | Crystal structure of botulinum neurotoxin serotype D/C VPI 5993 binding domain | | Descriptor: | GLYCEROL, Neurotoxin, SULFATE ION | | Authors: | Fu, Z, Karalewitz, A, Kroken, A, Baldwin, M.R, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Unique Ganglioside Binding Loop within Botulinum Neurotoxins C and D-SA .
Biochemistry, 49, 2010
|
|
7SN9
 
 | |
6FY4
 
 | |
8CZT
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
4YD2
 
 | | Nicked complex of human DNA Polymerase Mu with 2-nt gapped DNA substrate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*CP*GP*TP*AP*T)-3'), ... | | Authors: | Moon, A.F, Gosavi, R.A, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2015-02-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | Creative template-dependent synthesis by human polymerase mu.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5HI7
 
 | | Co-crystal structure of human SMYD3 with an aza-SAH compound | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl][3-(dimethylamino)propyl]amino}-5'-deoxyadenosine, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Elkins, P.A, Bonnette, W.G. | | Deposit date: | 2016-01-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of a Novel SMYD3 Inhibitor that Bridges the SAM-and MEKK2-Binding Pockets.
Structure, 24, 2016
|
|
6BY0
 
 | |
4YDD
 
 | | Crystal structure of the perchlorate reductase PcrAB from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tsai, C.-L, Youngblut, M.D, Tainer, J.A. | | Deposit date: | 2015-02-21 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
