5J8B
 
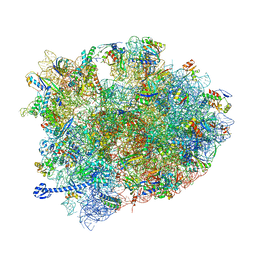 | | Crystal structure of Elongation Factor 4 (EF-4/LepA) in complex with GDPCP bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Lin, J, Steitz, T.A. | | Deposit date: | 2016-04-07 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Elongation factor 4 remodels the A-site tRNA on the ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8U1W
 
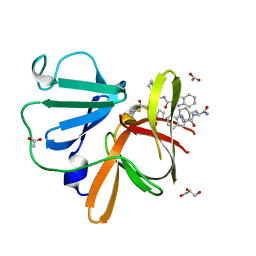 | | Structure of Norovirus (Hu/GII.4/Sydney/NSW0514/2012/AU) protease bound to inhibitor NV-004 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidase C37, ... | | Authors: | Eruera, A.R, Campbell, A.C, Krause, K.L. | | Deposit date: | 2023-09-03 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Inhibitor-Bound GII.4 Sydney 2012 Norovirus 3C-Like Protease.
Viruses, 15, 2023
|
|
8PI3
 
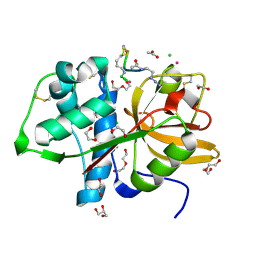 | | Cathepsin S Y132D mutant in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 2024
|
|
6VBN
 
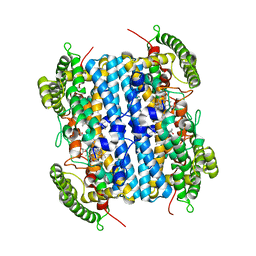 | | Crystal Structure of hTDO2 bound to inhibitor GNE1 | | Descriptor: | 1,5-anhydro-2,3-dideoxy-3-[(5S)-5H-imidazo[5,1-a]isoindol-5-yl]-D-threo-pentitol, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Harris, S.F, Oh, A. | | Deposit date: | 2019-12-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Implementation of the CYP Index for the Design of Selective Tryptophan-2,3-dioxygenase Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
5MVD
 
 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 1,5-dimethyl-3-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]pyrazole-4-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
6P7I
 
 | | Crystal structure of Human PRMT6 in complex with S-Adenosyl-L-Homocysteine and YS17-117 Compound | | Descriptor: | GLYCEROL, N-[3-(4-{[(2-aminoethyl)(methyl)amino]methyl}-1H-pyrrol-3-yl)phenyl]prop-2-enamide, N-[3-(4-{[(2-aminoethyl)(methyl)amino]methyl}-1H-pyrrol-3-yl)phenyl]propanamide, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a First-in-Class Protein Arginine Methyltransferase 6 (PRMT6) Covalent Inhibitor
J.Med.Chem., 63, 2020
|
|
5MXK
 
 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7G9. | | Descriptor: | 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR, ~{N}-(5-oxidanylidene-7,8-dihydro-6~{H}-naphthalen-2-yl)ethanamide | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MI3
 
 | | Structure of phosphorylated translation elongation factor EF-Tu from E. coli | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Talavera, A, Hendrix, J, Versees, W, De Gieter, S, Castro-Roa, D, Jurenas, D, Van Nerom, K, Vandenberk, N, Barth, A, De Greve, H, Hofkens, J, Zenkin, N, Loris, R, Garcia-Pino, A. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors.
Sci Adv, 4, 2018
|
|
8PE1
 
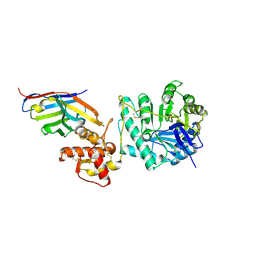 | | Crystal structure of Gel4 in complex with Nanobody 4 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 4, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6Y1I
 
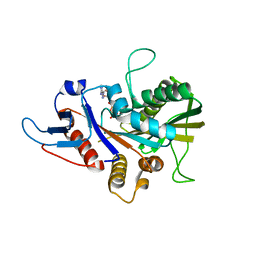 | |
6Y0L
 
 | | Crystal structure of human CD23 lectin domain N225D, K229E, S252N, T251N, R253G, S254G mutant | | Descriptor: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor membrane-bound form, SULFATE ION | | Authors: | Ilkow, V.F, Davies, A.M, Sutton, B.J, McDonnell, J.M. | | Deposit date: | 2020-02-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Reviving lost binding sites: Exploring calcium-binding site transitions between human and murine CD23.
Febs Open Bio, 11, 2021
|
|
6PBR
 
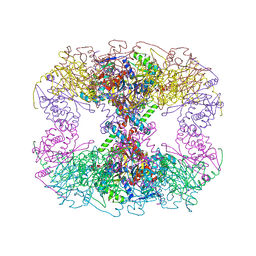 | | Catalytic domain of E.coli dihydrolipoamide succinyltransferase in I4 space group | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, SODIUM ION | | Authors: | Andi, B, Soares, A.S, Shi, W, Fuchs, M.R, McSweeney, S, Liu, Q. | | Deposit date: | 2019-06-14 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the dihydrolipoamide succinyltransferase catalytic domain from Escherichia coli in a novel crystal form: a tale of a common protein crystallization contaminant.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5MRR
 
 | | Crystal structure of L1 protease of Lysobacter sp. XL1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of L1 protease of Lysobacter sp. XL1
To Be Published
|
|
6PJJ
 
 | | Human PRPF4B bound to benzothiophene inhibitor 224 | | Descriptor: | 1,2-ETHANEDIOL, 4-(5-{[(3-aminophenyl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, PHOSPHATE ION, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-28 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | to be published
To Be Published
|
|
6VDP
 
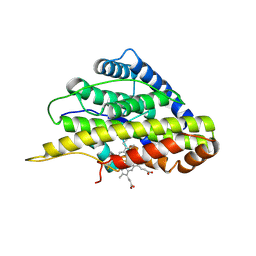 | | Crystal structure of SfmD truncated variant | | Descriptor: | 3-methyl-L-tyrosine peroxygenase, HEME C | | Authors: | Shin, I, Liu, A. | | Deposit date: | 2019-12-27 | | Release date: | 2021-03-10 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
6VDZ
 
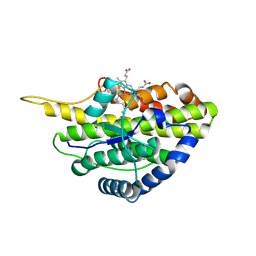 | | Crystal structure of reduced SfmD by soaking with sodium hydrosulfite | | Descriptor: | 3-methyl-L-tyrosine peroxygenase, HEME C | | Authors: | Shin, I, Liu, A. | | Deposit date: | 2019-12-27 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
5MSF
 
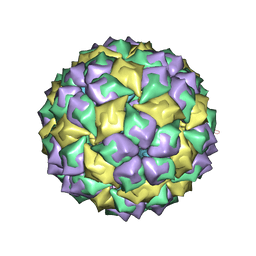 | | MS2 PROTEIN CAPSID/RNA COMPLEX | | Descriptor: | 5'-R(*CP*CP*GP*GP*AP*GP*GP*AP*UP*CP*AP*CP*CP*AP*CP*GP*GP*G)-3', MS2 PROTEIN CAPSID | | Authors: | Rowsell, S, Stonehouse, N.J, Convery, M.A, Adams, C.J, Ellington, A.D, Hirao, I, Peabody, D.S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 1998-05-15 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a series of RNA aptamers complexed to the same protein target.
Nat.Struct.Biol., 5, 1998
|
|
6VE0
 
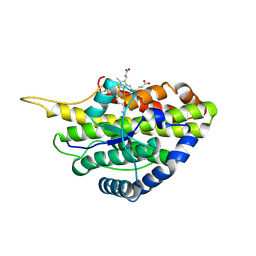 | | Crystal structure of reduced SfmD by soaking with sodium hydrosulfite | | Descriptor: | 3-methyl-L-tyrosine peroxygenase, HEME C | | Authors: | Shin, I, Liu, A. | | Deposit date: | 2019-12-27 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
5MUE
 
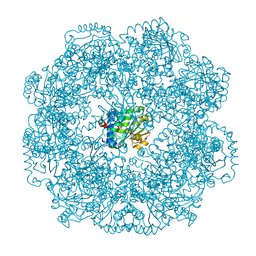 | | Self-assembled alpha-Tocopherol Transfer Protein Nanoparticles Promote Vitamin E Delivery Across an Endothelial Barrier | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, Alpha-tocopherol transfer protein, CHLORIDE ION, ... | | Authors: | Stocker, A, Aeschimann, W. | | Deposit date: | 2017-01-13 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Self-assembled alpha-Tocopherol Transfer Protein Nanoparticles Promote Vitamin E Delivery Across an Endothelial Barrier.
Sci Rep, 7, 2017
|
|
8POT
 
 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and the benzoxaborole cmpd9 in the editing conformation | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, [(1R,3'S,5S,6R,8R)-3'-(aminomethyl)-8-(6-aminopurin-9-yl)-4'-bromanyl-7'-[3-[methyl-(phenylmethyl)amino]propoxy]spiro[2,4,7-trioxa-3$l^{4}-borabicyclo[3.3.0]octane-3,1'-3H-2,1$l^{4}-benzoxaborole]-6-yl]methyl dihydrogen phosphate, ... | | Authors: | Palencia, A, Hoffmann, G. | | Deposit date: | 2023-07-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Targeting a microbiota Wolbachian aminoacyl-tRNA synthetase to block its pathogenic host.
Sci Adv, 10, 2024
|
|
6N6P
 
 | | Crystal structure of [FeFe]-hydrogenase in the presence of 7 mM Sodium dithionite | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Peters, J.W. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
8UFH
 
 | | Acinetobacter baylyi LptB2FG bound to Acinetobacter baylyi lipopolysaccharide and a macrocyclic peptide | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-2-[(2~{R},4~{R},5~{R},6~{R})-5-[(2~{S},4~{R},5~{R},6~{R})-4-[(2~{R},3~{R},4~{R},5~{S},6~{S})-3-acetamido-6-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-5-oxidanyl-oxan-2-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{S})-3-dodecanoyloxydodecanoyl]oxy-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-heptanoyloxynonanoyl]amino]-3-oxidanyl-4-[(3~{R})-3-oxidanyloctanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-5-[[(3~{S})-3-[(3~{R})-3-oxidanyldodecanoyl]oxydecanoyl]amino]-3-phosphonooxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, (7S,10S,13S,17P)-10-(4-aminobutyl)-7-(3-aminopropyl)-17-(6-aminopyridin-3-yl)-20-chloro-13-[(1H-indol-3-yl)methyl]-12-methyl-6,7,9,10,12,13,15,16-octahydropyrido[2,3-b][1,5,8,11,14]benzothiatetraazacycloheptadecine-8,11,14(5H)-trione, LPS export ABC transporter permease LptG, ... | | Authors: | Pahil, K.S, Gilman, M.S.A, Baidin, V, Kruse, A.C, Kahne, D. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A new antibiotic traps lipopolysaccharide in its intermembrane transporter.
Nature, 625, 2024
|
|
6VDQ
 
 | | Crystal structure of SfmD | | Descriptor: | 3-methyl-L-tyrosine peroxygenase, HEME C | | Authors: | Shin, I, Liu, A. | | Deposit date: | 2019-12-27 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
5MUL
 
 | | Glycoside Hydrolase BT3686 bound to Glucuronic Acid | | Descriptor: | Neuraminidase, beta-D-glucopyranuronic acid | | Authors: | Munoz-Munoz, J, Cartmell, A, Terrapon, N, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Unusual active site location and catalytic apparatus in a glycoside hydrolase family.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8PE2
 
 | | Crystal structure of Gel4 in complex with Nanobody 3 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 3, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
