5EYC
 
 | | Crystal structure of c-Met in complex with naphthyridinone inhibitor 5 | | Descriptor: | 6-[(1~{R})-1-[8-fluoranyl-6-(3-methyl-1,2-oxazol-5-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl]-1,6-naphthyridin-5-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of (R)-6-(1-(8-Fluoro-6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl)ethyl)-3-(2-methoxyethoxy)-1,6-naphthyridin-5(6H)-one (AMG 337), a Potent and Selective Inhibitor of MET with High Unbound Target Coverage and Robust In Vivo Antitumor Activity.
J.Med.Chem., 59, 2016
|
|
6UV2
 
 | | Crystal structure of the core domain of RNA helicase DDX17 with RNA pri-125a-oligo1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
5LNT
 
 | | Crystal structure of Arabidopsis thaliana Pdx1K166R-preI320 complex | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.1, [(~{E},4~{S})-4-azanyl-3-oxidanylidene-pent-1-enyl] dihydrogen phosphate | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2017-02-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
6UVM
 
 | | Cocrystal of BRD4(D1) with a methyl carbamate thiazepane inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[(7S)-7-(thiophen-2-yl)-6,7-dihydro-1,4-thiazepin-4(5H)-yl]ethan-1-one, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Pomerantz, W.C.K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains.
Acs Med.Chem.Lett., 10, 2019
|
|
5UMI
 
 | | Clostridium difficile TcdA-CROPs bound to PA50 Fab | | Descriptor: | PA50 Fab Heavy chain, PA50 Fab Light chain, Toxin A | | Authors: | Kroh, H.K, Chandrasekaran, R, Rosenthal, K, Woods, R, Jin, X, Ohi, M.D, Nyborg, A.C, Rainey, G.J, Warrener, P, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2017-01-27 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Use of a neutralizing antibody helps identify structural features critical for binding of Clostridium difficile toxin TcdA to the host cell surface.
J. Biol. Chem., 292, 2017
|
|
5LOY
 
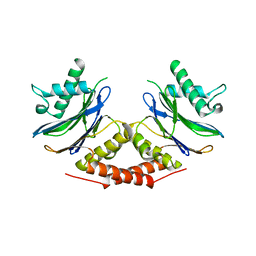 | |
5UP7
 
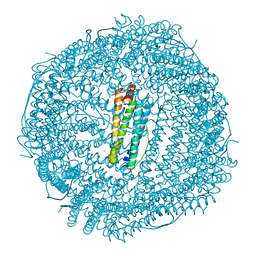 | | Crystal Structure of the Ni-bound Human Heavy-Chain Ferritin 122H-delta C-star variant | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bailey, J.B, Zhang, L, Chiong, J.A, Ahn, S, Tezcan, F.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Synthetic Modularity of Protein-Metal-Organic Frameworks.
J. Am. Chem. Soc., 139, 2017
|
|
6ERQ
 
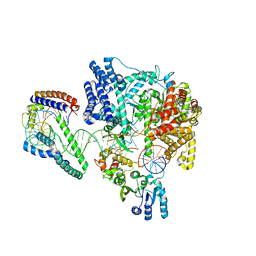 | | Structure of the human mitochondrial transcription initiation complex at the HSP promoter | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, Dimethyladenosine transferase 2, ... | | Authors: | Hillen, H.S, Morozov, Y.I, Sarfallah, A, Temiakov, D, Cramer, P. | | Deposit date: | 2017-10-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural Basis of Mitochondrial Transcription Initiation.
Cell, 171, 2017
|
|
5LR6
 
 | | Crystal Structure of COMT in complex with [3-(2,4-dimethyl-1,3-thiazol-5-yl)-1H-pyrazol-5-yl]-(4-phenylpiperazin-1-yl)methanone | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-18 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of COMT in complex with [3-(2,4-dimethyl-1,3-thiazol-5-yl)-1H-pyrazol-5-yl]-(4-phenylpiperazin-1-yl)methanone
To be published
|
|
8PBV
 
 | |
5OYM
 
 | | HIV Integrase Binding Domain of Lens Epithelium-Derived Growth Factor | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Rabbitts, T.H, Phillips, S.E.V, Cruz-Migoni, A, Carr, S.B, Hannon, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cloning, purification and structure determination of the HIV integrase-binding domain of lens epithelium-derived growth factor.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5F13
 
 | | Structure of Mn bound DUF89 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Nocek, B, Skarina, T, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | A family of metal-dependent phosphatases implicated in metabolite damage-control.
Nat.Chem.Biol., 12, 2016
|
|
6E7I
 
 | | Human ppGalNAcT2 I253A/L310A Mutant with EA2 and UDP | | Descriptor: | EA2, MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 2, ... | | Authors: | Bertozzi, C.R, Schumann, B, Agbay, A.J. | | Deposit date: | 2018-07-26 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bump-and-Hole Engineering Identifies Specific Substrates of Glycosyltransferases in Living Cells.
Mol.Cell, 78, 2020
|
|
5IPQ
 
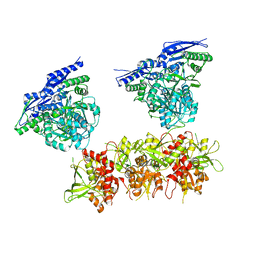 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 2 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
4QT7
 
 | |
5IST
 
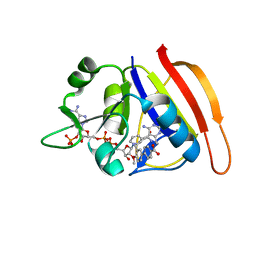 | | Staphylococcus aureus Dihydrofolate Reductase complexed with beta-NADPH, cyclic alpha-NADPH anomer and 3'-(3-(2,4-diamino-6-ethylpyrimidin-5-yl)prop-2-yn-1-yl)-4'-methoxy-[1,1'-biphenyl]-4-carboxylic acid (UCP1106) | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-4-methoxy-phenyl]benzoic acid, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Anderson, A.C, Reeve, S.M. | | Deposit date: | 2016-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.723 Å) | | Cite: | Charged Propargyl-Linked Antifolates Reveal Mechanisms of Antifolate Resistance and Inhibit Trimethoprim-Resistant MRSA Strains Possessing Clinically Relevant Mutations.
J. Med. Chem., 59, 2016
|
|
7KLJ
 
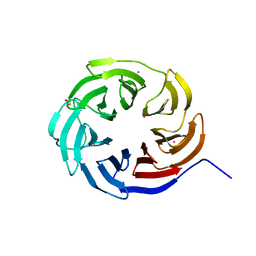 | | Crystal structure of the WD-repeat domain of human KIF21A | | Descriptor: | 1,2-ETHANEDIOL, Isoform 2 of Kinesin-like protein KIF21A, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Loppnau, P, Hutchinson, A, Seitova, A, Edwards, A.M, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-10-30 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of the WD-repeat domain of human KIF21A
To be Published
|
|
6ESB
 
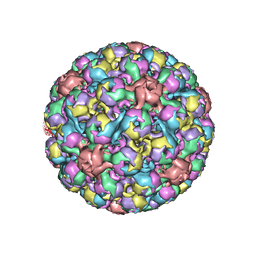 | | BK polyomavirus + 20 mM GT1b oligosaccharide | | Descriptor: | CALCIUM ION, Capsid protein VP1, Minor capsid protein VP2, ... | | Authors: | Hurdiss, D.L, Ranson, N.A. | | Deposit date: | 2017-10-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Structure of an Infectious Human Polyomavirus and Its Interactions with Cellular Receptors.
Structure, 26, 2018
|
|
6V1O
 
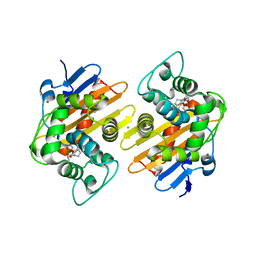 | | Structure of OXA-48 bound to QPX7728 at 1.80 A | | Descriptor: | (1aR,7bS)-5-fluoro-2-hydroxy-1,1a,2,7b-tetrahydrocyclopropa[c][1,2]benzoxaborinine-4-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Boronic Acid QPX7728, an Ultrabroad-Spectrum Inhibitor of Serine and Metallo-beta-lactamases.
J.Med.Chem., 63, 2020
|
|
5LQJ
 
 | | Crystal Structure of COMT in complex with 3-cyclopropyl-5-methyl-4-phenyl-1,2,4-triazole | | Descriptor: | 3-cyclopropyl-5-methyl-4-phenyl-1,2,4-triazole, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-17 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of COMT in complex with 3-cyclopropyl-5-methyl-4-phenyl-1,2,4-triazole
To be published
|
|
6SFE
 
 | | CRYSTAL STRUCTURE OF DHQ1 FROM SALMONELLA TYPHI COVALENTLY MODIFIED BY COMPOUND 7 | | Descriptor: | (1~{S},3~{S},4~{S},5~{R})-3-(aminomethyl)-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
6BZU
 
 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody 19B3 | | Descriptor: | 19B3 Heavy Chain, 19B3 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BZY
 
 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the 22D11 broadly neutralizing antibody | | Descriptor: | 22D11 Heavy Chain, 22D11 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5BQM
 
 | | Crystal structure of SXN101959, a Clostridium botulinum neurotoxin type D derivative and targeted secretion inhibitor | | Descriptor: | Botulinum neurotoxin type D, Somatoliberin,Botulinum neurotoxin type D, ZINC ION | | Authors: | Masuyer, G, Davies, J.R, Moore, K, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of Clostridium botulinum neurotoxin type D as a platform for the development of targeted secretion inhibitors.
Sci Rep, 5, 2015
|
|
5UVJ
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
