7OEJ
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ090 | | Descriptor: | 4-[[(3S)-3-cyclopropyl-2-azaspiro[3.3]heptan-2-yl]methyl]-N-[[(3R)-1-[6-(methylamino)pyrimidin-4-yl]-3-oxidanyl-piperidin-3-yl]methyl]-2-oxidanyl-benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
5D4Q
 
 | | Crystal structure of GASDALIE IgG1 Fc | | Descriptor: | Ig gamma-1 chain C region, alpha-L-fucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ahmed, A.A, Bjorkman, P.J. | | Deposit date: | 2015-08-08 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural characterization of GASDALIE Fc bound to the activating Fc receptor Fc gamma RIIIa.
J.Struct.Biol., 194, 2016
|
|
7OEH
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ059b | | Descriptor: | 4-[[(3S)-3-cyclopropyl-2-azaspiro[3.3]heptan-2-yl]methyl]-N-[[(3S)-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
1K1R
 
 | | HETERODUPLEX OF CHIRALLY PURE R-METHYLPHOSPHONATE/DNA DUPLEX | | Descriptor: | 5'-D(*CP*(CMR)P*(RMP)P*(RMP)P*(RMP)P*(CMR)P*(RMP))-3', 5'-D(*TP*GP*TP*TP*TP*GP*GP*C)-3' | | Authors: | Thiviyanathan, V, Vyazovkina, K.V, Gozansky, E.K, Bichenkova, E, Abramova, T.V, Luxon, B.A, Lebedev, A.V, Gorenstein, D.G. | | Deposit date: | 2001-09-25 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry.
Biochemistry, 41, 2002
|
|
7OEM
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ120 | | Descriptor: | 4-[(4,4-dimethylpiperidin-1-yl)methyl]-2-methylsulfanyl-~{N}-[[(3~{R})-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
5UWY
 
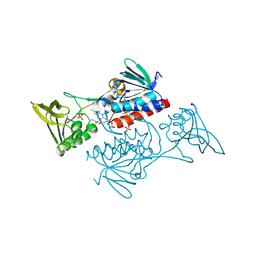 | | The crystal structure of thioredoxin reductase from Streptococcus pyogenes MGAS5005 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, Thioredoxin reductase | | Authors: | Tan, K, Zhou, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-21 | | Release date: | 2017-03-15 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of thioredoxin reductase from Streptococcus pyogenes MGAS5005
To Be Published
|
|
4IB8
 
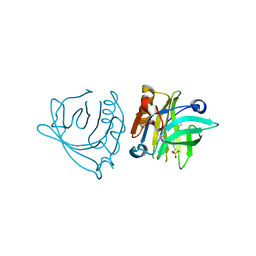 | | Bovine beta-lactoglobulin (isoform A) in complex with dodecyl sulphate (SDS) | | Descriptor: | DODECYL SULFATE, beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Swiatek, S, Dziedzicka-Wasylewska, M, Lewinski, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The differences in binding 12-carbon aliphatic ligands by bovine beta-lactoglobulin isoform A and B studied by isothermal titration calorimetry and X-ray crystallography
J.Mol.Recognit., 26, 2013
|
|
1K1H
 
 | | HETERODUPLEX OF CHIRALLY PURE METHYLPHOSPHONATE/DNA DUPLEX | | Descriptor: | 5'-D(*CP*(CMR)P*(RMP)P*(RMP)P*(SMP)P*(CMR)P*(RMP))-3', 5'-D(*TP*GP*TP*TP*TP*GP*GP*C)-3' | | Authors: | Thiviyanathan, V, Vyazovkina, K.V, Gozansky, E.K, Bichenchova, E, Abramova, T.V, Luxon, B.A, Lebedev, A.V, Gorenstein, D.G. | | Deposit date: | 2001-09-25 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry.
Biochemistry, 41, 2002
|
|
4IBV
 
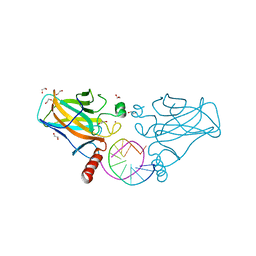 | | Human p53 core domain with hot spot mutation R273C and second-site suppressor mutation S240R in sequence-specific complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA (5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3'), ... | | Authors: | Eldar, A, Rozenberg, H, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2012-12-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
6WAN
 
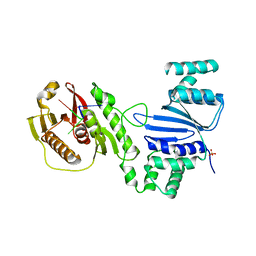 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 3'3'3'-cAAA | | Descriptor: | Cyclic RNA (R(P*AP*AP*A), SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
2VUZ
 
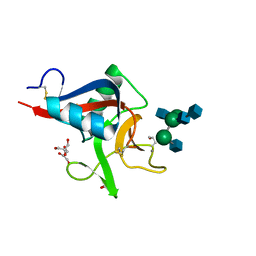 | | Crystal structure of Codakine in complex with biantennary nonasaccharide at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CODAKINE, ... | | Authors: | Gourdine, J.P, Cioci, G.C, Miguet, L, Unverzagt, C, Varrot, A, Gauthier, C, Smith-Ravin, E.J, Imberty, A. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High Affinity Interaction between a Bivalve C-Type Lectin and a Biantennary Complex-Type N-Glycan Revealed by Crystallography and Microcalorimetry.
J.Biol.Chem., 283, 2008
|
|
4IBF
 
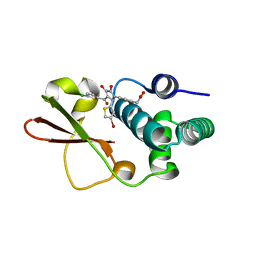 | | Ebola virus VP35 bound to small molecule | | Descriptor: | (4-{(2R)-2-(4-bromothiophen-2-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}phenyl)acetic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
5DD4
 
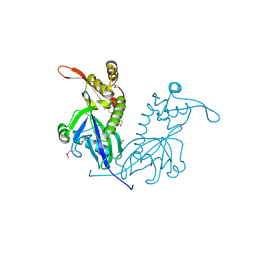 | | Apo structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, TRANSCRIPTIONAL REGULATOR AraR | | Authors: | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
7O2P
 
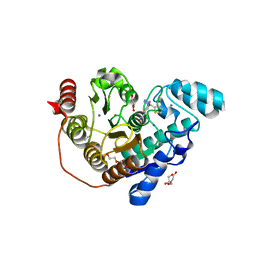 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with ITF3756 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zrubek, K, Sandrone, G, Cukier, C.D, Stevenazzi, A. | | Deposit date: | 2021-03-31 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Fluorination in the Histone Deacetylase 6 (HDAC6) Selectivity of Benzohydroxamate-Based Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7ALV
 
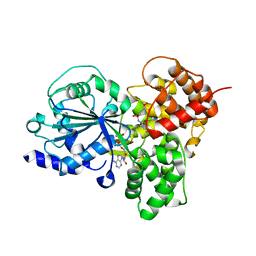 | | Crystal Structure of NLRP3 NACHT domain in complex with a potent inhibitor | | Descriptor: | 1-[4-chloranyl-2,6-di(propan-2-yl)phenyl]-3-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Dekker, C, Hinniger, A. | | Deposit date: | 2020-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.835 Å) | | Cite: | Crystal Structure of NLRP3 NACHT Domain With an Inhibitor Defines Mechanism of Inflammasome Inhibition.
J.Mol.Biol., 433, 2021
|
|
5UQ4
 
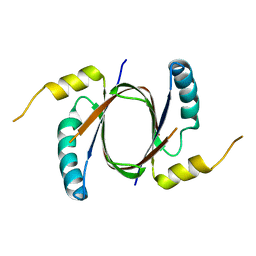 | | Crystal structure of Heme-Degrading Protein Rv3592 from Mycobacterium tuberculosis - heme free with cleaved protein | | Descriptor: | Monooxygenase | | Authors: | Chang, C, Chhor, G, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-06 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal structure of Heme-Degrading Protein Rv3592 from Mycobacterium tuberculosis - heme free with cleaved protein.
To Be Published
|
|
7B7G
 
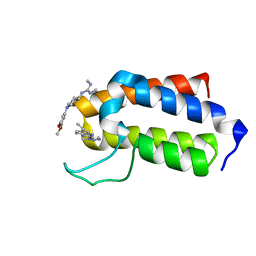 | | BAZ2A bromodomain in complex with compounds MS04 and B11 | | Descriptor: | 1-[1-(3,5-dimethoxyphenyl)piperidin-4-yl]-2,3-dimethyl-guanidine, 5-ethyl-2-(3-methyl-1,2,3-triazol-4-yl)-1~{H}-benzimidazole, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.428 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
7ALY
 
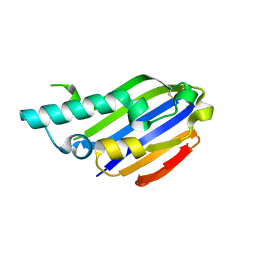 | |
7AMX
 
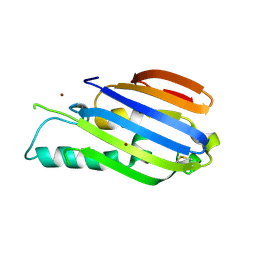 | |
7B82
 
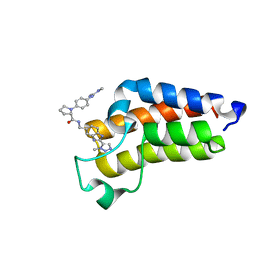 | | BAZ2A bromodomain in complex with triazole compound MS04-TN03 | | Descriptor: | (2~{S})-1-[4-[[(~{E})-~{N},~{N}'-dimethylcarbamimidoyl]amino]phenyl]-~{N}-[[2-(3-methyl-1,2,3-triazol-4-yl)-1~{H}-benzimidazol-5-yl]methyl]pyrrolidine-2-carboxamide, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
6PIY
 
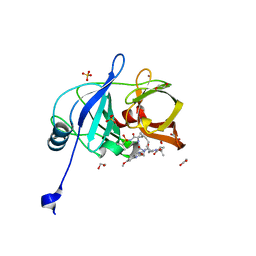 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-2 (NR02-61) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
3WZP
 
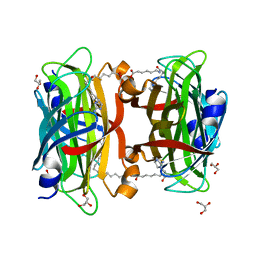 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.2 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, GLYCEROL, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
2WF3
 
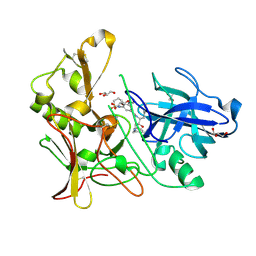 | | Human BACE-1 in complex with 6-(ethylamino)-N-((1S,2R)-2-hydroxy-3-(((3-(methyloxy)phenyl)methyl)amino)-1-(phenylmethyl)propyl)-1-methyl-1, 3,4,5-tetrahydro-2,1-benzothiazepine-8-carboxamide 2,2-dioxide | | Descriptor: | BETA-SECRETASE 1, GLYCEROL, N-{(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-6-(ETHYLAMINO)-1-METHYL-1,3,4,5-TETRAHYDRO-2,1-BENZOTHIAZEPINE-8-CARBOXAMIDE 2,2-DIOXIDE | | Authors: | Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-19 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Second Generation of Bace-1 Inhibitors Part 2: Optimisation of the Non-Prime Side Substituent.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1JII
 
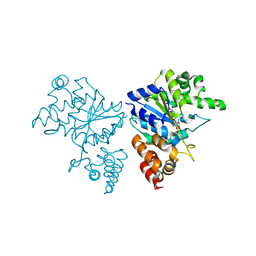 | | Crystal structure of S. aureus TyrRS in complex with SB-219383 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]- (2,4,5,8-TETRAHYDROXY-7-OXA-2-AZA-BICYCLO[3.2.1]OCT-3-YL)- ACETIC ACID, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
5DHV
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-22 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
