6VTI
 
 | | Solution NMR structure of the N-terminal domain of the Serine/threonine-protein phosphatase 1 regulatory subunit 10, PPP1R10 | | Descriptor: | Serine/threonine-protein phosphatase 1 regulatory subunit 10 | | Authors: | Lemak, A, Wei, Y, Duan, S, Houliston, S, Penn, L.Z, Arrowsmith, C.H. | | Deposit date: | 2020-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase.
Nucleic Acids Res., 50, 2022
|
|
6VVQ
 
 | |
6ZA0
 
 | | Structure of the transcriptional repressor Atu1419 (VanR) in complex with a fortuitous citrate from agrobacterium fabrum (P21212 space group) | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
6ZA3
 
 | | Structure of the transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum in complex a palindromic DNA (C2221 space group) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
8ERM
 
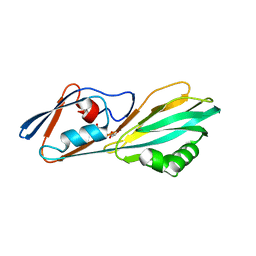 | | Crystal structure of FliC D2/D3 domains from Pseudomonas aeruginosa PAO1 | | Descriptor: | B-type flagellin, GLYCEROL, SULFATE ION | | Authors: | Nedeljkovic, M, Bonsor, D.A, Postel, S, Sundberg, E.J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.475 Å) | | Cite: | An unbroken network of interactions connecting flagellin domains is required for motility in viscous environments.
Plos Pathog., 19, 2023
|
|
6VX8
 
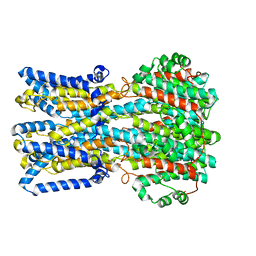 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PTQ
 
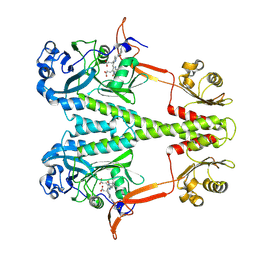 | | Dark, Room Temperature, PCM Myxobacterial Phytochrome, P2, Wild Type | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, BENZAMIDINE, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
5NME
 
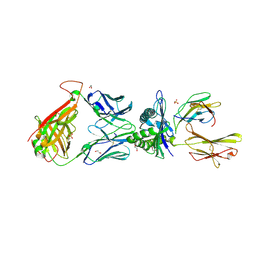 | | 868 TCR in complex with HLA A02 presenting SLYNTVATL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Dual Molecular Mechanisms Govern Escape at Immunodominant HLA A2-Restricted HIV Epitope.
Front Immunol, 8, 2017
|
|
6VX9
 
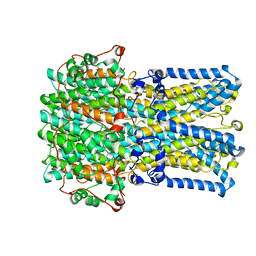 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZEJ
 
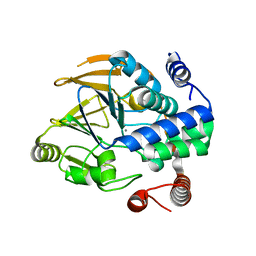 | | Structure of PP1-Phactr1 chimera [PP1(7-304) + linker (SGSGS) + Phactr1(526-580)] | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
7NL6
 
 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | CALCIUM ION, DC-SIGN, CRD domain, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
6VYN
 
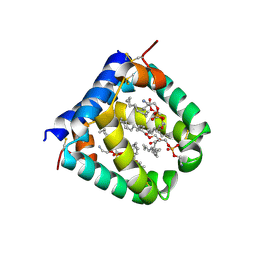 | | N-terminal domain of mouse surfactant protein B with bound lipid, wild type | | Descriptor: | (7Z,19R,22R)-25-amino-22-hydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphapentacos-7-en-19-yl (9Z)-octadec-9-enoate, Pulmonary surfactant-associated protein B | | Authors: | Rapoport, T.A, Bodnar, N.O. | | Deposit date: | 2020-02-27 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Lamellar Body Formation by Lung Surfactant Protein B.
Mol.Cell, 81, 2021
|
|
8B37
 
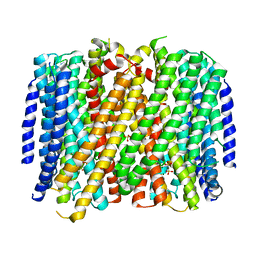 | | Crystal structure of Pyrobaculum aerophilum potassium-independent proton pumping membrane integral pyrophosphatase in complex with imidodiphosphate and magnesium, and with bound sulphate | | Descriptor: | IMIDODIPHOSPHORIC ACID, K(+)-insensitive pyrophosphate-energized proton pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Wilkinson, C, Vidilaseris, K, Ribeiro, O, Liu, J, Hillier, J, Malinen, A, Gehl, B, Jeuken, L.C, Pearson, A.R, Goldman, A. | | Deposit date: | 2022-09-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8EOI
 
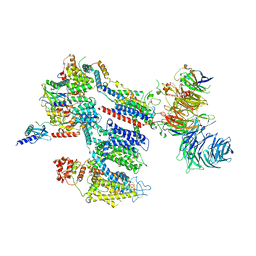 | | Structure of a human EMC:human Cav1.2 channel complex in GDN detergent | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Mondal, A, Abderemane-Ali, F, Minor, D.L. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | EMC chaperone-Ca V structure reveals an ion channel assembly intermediate.
Nature, 619, 2023
|
|
6W1B
 
 | |
8F2R
 
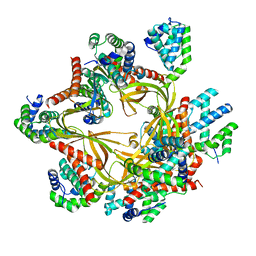 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
5NBJ
 
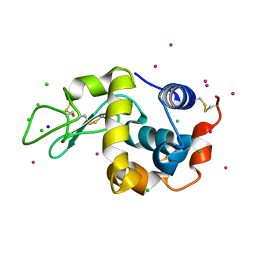 | | DLS Tetragonal - ReHEWL | | Descriptor: | CHLORIDE ION, Lysozyme C, RHENIUM, ... | | Authors: | Brink, A, Helliwell, J.R. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.266 Å) | | Cite: | New leads for fragment-based design of rhenium/technetium radiopharmaceutical agents.
IUCrJ, 4, 2017
|
|
6W3S
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-leucine | | Descriptor: | GLYCEROL, LEUCINE, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
8EIT
 
 | | Structure of FFAR1-Gq complex bound to DHA | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EJC
 
 | | Structure of FFAR1-Gq complex bound to TAK-875 | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, Free fatty acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6W3V
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-phenylalanine | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein, PHENYLALANINE, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6Q1M
 
 | | Crystal structure of the wheat dwarf virus Rep domain | | Descriptor: | GLYCEROL, Replication-associated protein | | Authors: | Litzau, L.A, Everett, B.A, Evans III, R.L, Shi, K, Tompkins, K, Gordon, W.R. | | Deposit date: | 2019-08-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of the Wheat dwarf virus Rep domain.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8BIK
 
 | | Crystal structure of human AMPK heterotrimer in complex with allosteric activator C455 | | Descriptor: | (3~{R},3~{a}~{R},6~{R},6~{a}~{R})-6-[[6-chloranyl-5-[4-[4-[[dimethyl(oxidanyl)-$l^{4}-sulfanyl]amino]phenyl]phenyl]-3~{H}-imidazo[4,5-b]pyridin-2-yl]oxy]-2,3,3~{a},5,6,6~{a}-hexahydrofuro[3,2-b]furan-3-ol, 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Schimpl, M, Mather, K.M, Boland, M.L, Rivers, E.L, Srivastava, A, Hemsley, P, Robinson, J, Wan, P.T, Hansen, J, Read, J.A, Trevaskis, J.L, Smith, D.M. | | Deposit date: | 2022-11-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Direct beta 1/ beta 2 AMPK activation reduces liver steatosis but not fibrosis in a mouse model of non-alcoholic steatohepatitis
Biorxiv, 2024
|
|
6QA8
 
 | | Glycogen Phosphorylase b in complex with 28 | | Descriptor: | (5~{S},7~{R},8~{S},9~{S},10~{R})-7-(hydroxymethyl)-8,9,10-tris(oxidanyl)-2-phenyl-6-oxa-1,3-diazaspiro[4.5]dec-1-en-4-one, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Glucopyranosylidene-spiro-imidazolinones, a New Ring System: Synthesis and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetics and X-ray Crystallography.
J.Med.Chem., 62, 2019
|
|
8BRH
 
 | | Co-crystal structure of She4 with Myo4 peptide | | Descriptor: | KLLA0E16699p, Myo4 peptide (LYS-PHE-ILE-VAL-SER-HIS-TYR) | | Authors: | Arnese, R, Gudino, R, Meinhart, A, Clausen, T. | | Deposit date: | 2022-11-23 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | UNC-45 assisted myosin folding depends on a conserved FX 3 HY motif implicated in Freeman Sheldon Syndrome.
Nat Commun, 15, 2024
|
|
