6VOU
 
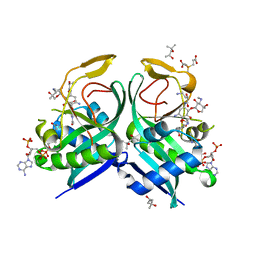 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with acetylated-plazomicin and CoA | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-{[(2S,3R)-3-(acetylamino)-6-{[(2-hydroxyethyl)amino]methyl}-3,4-dihydro-2H-pyran-2-yl]oxy}-5-amino-2-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-3-hydroxycyclohexyl]-4-amino-2-hydroxybutanamide, (4S)-2-METHYL-2,4-PENTANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, ... | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-01-31 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for plazomicin antibiotic action and resistance.
Commun Biol, 4, 2021
|
|
6VR2
 
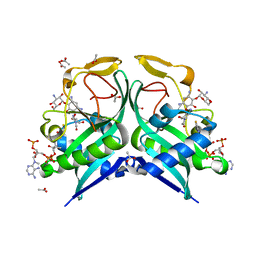 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with acetylated-tobramycin and CoA | | Descriptor: | (1S,2S,3R,4S,6R)-3-{[2-(acetylamino)-6-amino-2,3,6-trideoxy-alpha-D-ribo-hexopyranosyl]oxy}-4,6-diamino-2-hydroxycycloh exyl 3-amino-3-deoxy-alpha-D-glucopyranoside, ACETATE ION, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-02-06 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
8CIE
 
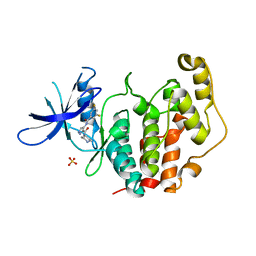 | | Crystal structure of the human CDKL5 kinase domain with compound YL-354 | | Descriptor: | 4-[[3,5-bis(fluoranyl)phenyl]carbonylamino]-~{N}-piperidin-4-yl-1~{H}-pyrazole-3-carboxamide, Cyclin-dependent kinase-like 5, SULFATE ION | | Authors: | Richardson, W, Chen, X, Newman, J.A, Bakshi, S, Lakshminarayana, B, Brooke, L, Bullock, A.N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective CDKL5/GSK3 Chemical Probe That Is Neuroprotective.
Acs Chem Neurosci, 14, 2023
|
|
5NA9
 
 | |
7Q51
 
 | | yeast Gid10 bound to a Phe/N-peptide | | Descriptor: | CHLORIDE ION, FWLPANLW peptide, Uncharacterized protein YGR066C | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
7Q50
 
 | | human Gid4 bound to a Phe/N-peptide | | Descriptor: | FDVSWFMG peptide, Glucose-induced degradation protein 4 homolog | | Authors: | Chrustowicz, J, Sherpa, D, Loke, M.S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
6WA1
 
 | |
5MZP
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with caffeine at 2.1A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CAFFEINE, ... | | Authors: | Cheng, K.Y.R, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
6Z9L
 
 | | Enterococcal PrgA | | Descriptor: | Poly-alanine peptide, PrgA, SULFATE ION | | Authors: | Berntsson, R.P.A, Schmitt, A. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.063 Å) | | Cite: | Enterococcal PrgA Extends Far Outside the Cell and Provides Surface Exclusion to Protect against Unwanted Conjugation.
J.Mol.Biol., 432, 2020
|
|
7BNN
 
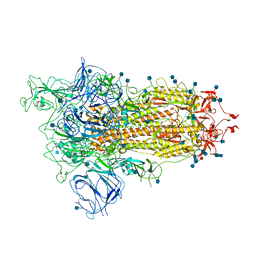 | | Open conformation of D614G SARS-CoV-2 spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BTE
 
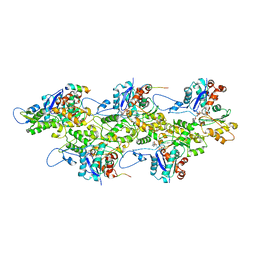 | | Lifeact-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kumari, A, Ragunath, V.K, Sirajuddin, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into actin filament recognition by commonly used cellular actin markers.
Embo J., 39, 2020
|
|
6WEJ
 
 | | Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZE8
 
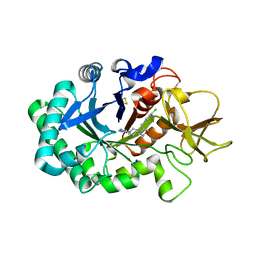 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound OATD-01 | | Descriptor: | 5-(4-((2S,5S)-5-(4-chlorobenzyl)-2-methylmorpholino)piperidin-1-yl)-4H-1,2,4-triazol-3-amine, Chitotriosidase-1, GLYCEROL, ... | | Authors: | Nowotny, M, Bartlomiejczak, A, Napiorkowska-Gromadzka, A. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of OATD-01 , a First-in-Class Chitinase Inhibitor as Potential New Therapeutics for Idiopathic Pulmonary Fibrosis.
J.Med.Chem., 63, 2020
|
|
6ZGC
 
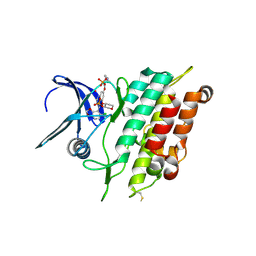 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Saracatinib (AZD0530) | | Descriptor: | Activin receptor type I, N-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-7-[2-(4-METHYLPIPERAZIN-1-YL)ETHOXY]-5-(TETRAHYDRO-2H-PYRAN-4-YLOXY)QUINAZOLIN-4-AMINE, PHOSPHATE ION, ... | | Authors: | Williams, E.P, Galan Bartual, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Saracatinib is an efficacious clinical candidate for fibrodysplasia ossificans progressiva.
JCI Insight, 6, 2021
|
|
6WGD
 
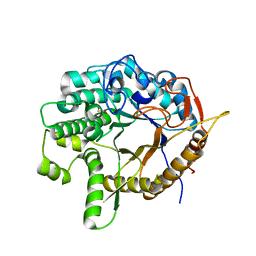 | | Crystal structure of a 6-phospho-beta-glucosidase from Bacillus licheniformis | | Descriptor: | 1,2-ETHANEDIOL, 6-phospho-beta-glucosidase | | Authors: | Liberato, M.V, Popov, A, Polikarpov, I. | | Deposit date: | 2020-04-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Structure, Bioinformatics Analysis, and Substrate Specificity of a 6-Phospho-beta-glucosidase Glycoside Hydrolase 1 Enzyme from Bacillus licheniformis .
J.Chem.Inf.Model., 60, 2020
|
|
6QLD
 
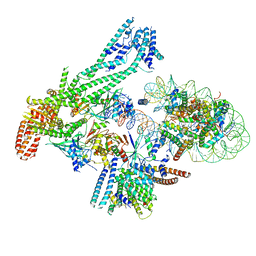 | | Structure of inner kinetochore CCAN-Cenp-A complex | | Descriptor: | DNA (125-MER), Histone H2A.1, Histone H2B.1, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
7BS8
 
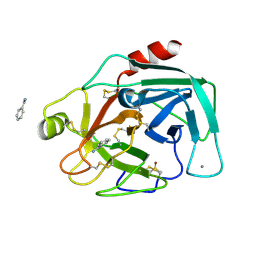 | | Bovine Pancreatic Trypsin with Benzamidine (Cryo) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
6Q2M
 
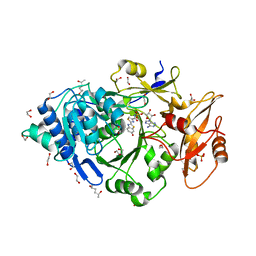 | | Crystal structure of Photinus pyralis Luciferase Pps6 mutant in complex with DLSA | | Descriptor: | (2S,5S)-hexane-2,5-diol, 1,2-ETHANEDIOL, 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, ... | | Authors: | Patel, K.D, Gulick, A.M. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mutagenesis and Structural Studies Reveal the Basis for the Activity and Stability Properties That Distinguish thePhotinusLuciferasesscintillansandpyralis.
Biochemistry, 58, 2019
|
|
6Z55
 
 | | Crystal structure of CLK3 in complex with macrocycle ODS2004070 | | Descriptor: | 1,2-ETHANEDIOL, 7,10-Dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.12,6.017,20]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxylic acid, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2004070
To Be Published
|
|
7MZQ
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with fucose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, beta-L-fucopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
6WEK
 
 | | Structure of cGMP-bound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8EYH
 
 | |
7MZO
 
 | | Crystal structure of the UcaD lectin-binding domain | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZS
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with galactose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, alpha-D-galactopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZP
 
 | | Crystal structure of the UclD lectin-binding domain | | Descriptor: | F17-like fimbril adhesin subunit UclD, IODIDE ION | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
