8QBN
 
 | |
5I7C
 
 | | Centrosomin-motif 2 (CM2) domain of Drosophila melanogaster Centrosomin (Cnn) | | 分子名称: | Centrosomin, ZINC ION | | 著者 | Feng, Z, Cottee, M.A, Johnson, S, Lea, S.M. | | 登録日 | 2016-02-17 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.804 Å) | | 主引用文献 | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
6PA7
 
 | | The cryo-EM structure of the human DNMT3A2-DNMT3B3 complex bound to nucleosome. | | 分子名称: | CHLORIDE ION, DNA (167-MER), DNA (cytosine-5)-methyltransferase 3A, ... | | 著者 | Xu, T.H, Liu, M, Zhou, X.E, Liang, G, Zhao, G, Xu, H.E, Melcher, K, Jones, P.A. | | 登録日 | 2019-06-11 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B.
Nature, 586, 2020
|
|
7RM5
 
 | | MicroED structure of the human adenosine receptor at 2.8A | | 分子名称: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, CHOLESTEROL, ... | | 著者 | Martynowycz, M.W, Shiriaeva, A, Ge, X, Hattne, J, Nannenga, B.L, Cherezov, V, Gonen, T. | | 登録日 | 2021-07-26 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.79 Å) | | 主引用文献 | MicroED structure of the human adenosine receptor determined from a single nanocrystal in LCP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3LHH
 
 | | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1. | | 分子名称: | ADENOSINE MONOPHOSPHATE, CBS domain protein | | 著者 | Tan, K, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-01-22 | | 公開日 | 2010-02-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1.
To be Published
|
|
1A5Q
 
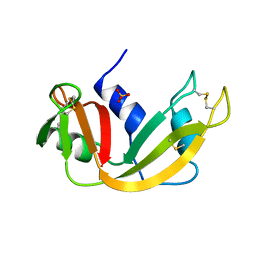 | | P93A VARIANT OF BOVINE PANCREATIC RIBONUCLEASE A | | 分子名称: | RIBONUCLEASE A, SULFATE ION | | 著者 | Pearson, M.A, Karplus, P.A, Dodge, R.W, Laity, J.H, Scheraga, H.A. | | 登録日 | 1998-02-17 | | 公開日 | 1998-05-27 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of two mutants that have implications for the folding of bovine pancreatic ribonuclease A.
Protein Sci., 7, 1998
|
|
6RMK
 
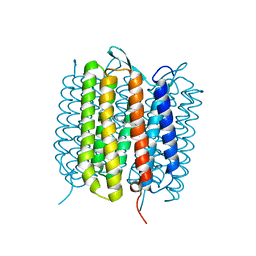 | | Bacteriorhodopsin, dark state, cell 2, refined using the same protocol as sub-ps time delays | | 分子名称: | Bacteriorhodopsin, RETINAL | | 著者 | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | 登録日 | 2019-05-07 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
3LIR
 
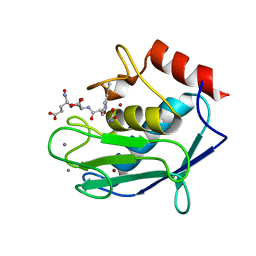 | | Human MMP12 in complex with non-zinc chelating inhibitor | | 分子名称: | CALCIUM ION, GLYCINE, Macrophage metalloelastase, ... | | 著者 | Stura, E.A, Dive, V, Devel, L, Czarny, B, Vera, L, Beau, F. | | 登録日 | 2010-01-25 | | 公開日 | 2010-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Insights from selective non-phosphinic inhibitors of MMP-12 tailored to fit with an S1' loop canonical conformation.
J.Biol.Chem., 285, 2010
|
|
4QU8
 
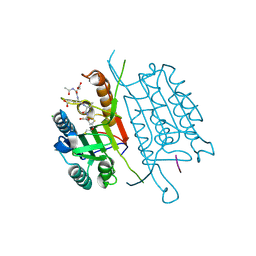 | | Caspase-3 M61A V266H | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CHLORIDE ION, ... | | 著者 | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | 登録日 | 2014-07-10 | | 公開日 | 2014-11-05 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.715 Å) | | 主引用文献 | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
7RAH
 
 | |
1AES
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (IMIDAZOLE) | | 分子名称: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | 登録日 | 1997-02-25 | | 公開日 | 1997-09-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
5IBZ
 
 | | Crystal structure of a novel cyclase (pfam04199). | | 分子名称: | ACETYLPHOSPHATE, TRIETHYLENE GLYCOL, Uncharacterized protein | | 著者 | Nocek, B, Skarina, T, Brown, G, Joachimiak, A, Savchenko, A, Yakunin, A. | | 登録日 | 2016-02-22 | | 公開日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.611 Å) | | 主引用文献 | Crystal structure of a novel cyclase (pfam04199).
To Be Published
|
|
4XPO
 
 | | Crystal structure of a novel alpha-galactosidase from Pedobacter saltans | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-glucosidase | | 著者 | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | 登録日 | 2015-01-17 | | 公開日 | 2015-05-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
6E5S
 
 | |
3LLK
 
 | | Sulfhydryl Oxidase Fragment of Human QSOX1 | | 分子名称: | CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, Sulfhydryl oxidase 1 | | 著者 | Alon, A, Fass, D. | | 登録日 | 2010-01-29 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | QSOX contains a pseudo-dimer of functional and degenerate sulfhydryl oxidase domains.
Febs Lett., 584, 2010
|
|
5G0Y
 
 | |
4XQ5
 
 | | Human-infecting H10N8 influenza virus retains strong preference for avian-type receptors | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | 著者 | Tzarum, N, Zhang, H, Zhu, X, Wilson, I.A. | | 登録日 | 2015-01-19 | | 公開日 | 2015-04-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.592 Å) | | 主引用文献 | A Human-Infecting H10N8 Influenza Virus Retains a Strong Preference for Avian-type Receptors.
Cell Host Microbe, 17, 2015
|
|
4XR0
 
 | |
8R5K
 
 | |
5IEW
 
 | |
6P05
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with compound 27 | | 分子名称: | Bromodomain-containing protein 4, GLYCEROL, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(1-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-3-yl)-1H-indol-4-yl}ethanesulfonamide | | 著者 | Ratia, K.M, Xiong, R, Li, Y, Zhao, J, Gutgesell, L.M, Shen, Z, Dye, K, Dubrovyskyii, O, Zhao, H, Huang, F, Tonetti, D.A, Thatcher, G.R. | | 登録日 | 2019-05-16 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
6S77
 
 | | Crystal structure of CARM1 N265Y mutant in complex with inhibitor AA183 | | 分子名称: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-(pyridin-2-ylamino)propyl]amino]-2-azanyl-butanoic acid, Histone-arginine methyltransferase CARM1 | | 著者 | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | 登録日 | 2019-07-04 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
4XSX
 
 | |
6S7Y
 
 | |
4QU0
 
 | | Caspase-3 Y195AV266H | | 分子名称: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, AZIDE ION, Caspase-3 | | 著者 | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | 登録日 | 2014-07-10 | | 公開日 | 2014-11-05 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.954 Å) | | 主引用文献 | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
