6J5D
 
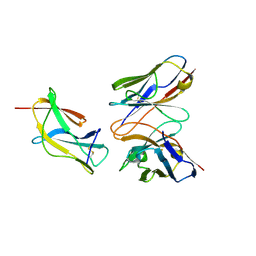 | | Complex structure of MAb 4.2-scFv with louping ill virus envelope protein Domain III | | 分子名称: | Envelope, antibody heavy chain, antibody light chain | | 著者 | Yang, X, Qi, J, Peng, R, Dai, L, Gould, E.A, Tien, P, Gao, G.F. | | 登録日 | 2019-01-10 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular Basis of a Protective/Neutralizing Monoclonal Antibody Targeting Envelope Proteins of both Tick-Borne Encephalitis Virus and Louping Ill Virus.
J. Virol., 93, 2019
|
|
1T3M
 
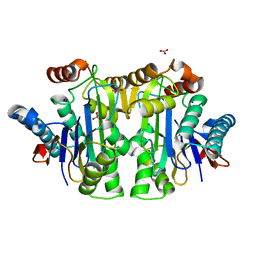 | | Structure of the isoaspartyl peptidase with L-asparaginase activity from E. coli | | 分子名称: | NITRATE ION, Putative L-asparaginase, SODIUM ION | | 著者 | Prahl, A, Pazgier, M, Hejazi, M, Lockau, W, Lubkowski, J. | | 登録日 | 2004-04-27 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure of the isoaspartyl peptidase with L-asparaginase activity from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T2S
 
 | | Structural basis for 3' end recognition of nucleic acids by the Drosophila Argonaute 2 PAZ domain | | 分子名称: | 5'-D(*CP*TP*CP*AP*C)-3', Argonaute 2 | | 著者 | Lingel, A, Simon, B, Izaurralde, E, Sattler, M. | | 登録日 | 2004-04-22 | | 公開日 | 2004-06-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nucleic acid 3'-end recognition by the Argonaute2 PAZ domain.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4RJ2
 
 | |
7TEA
 
 | | Crystal structure of S. aureus GlnR-DNA complex | | 分子名称: | CALCIUM ION, DNA (5'-D(*CP*GP*TP*GP*TP*CP*AP*GP*AP*TP*AP*AP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*GP*TP*CP*AP*GP*AP*TP*TP*AP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), ... | | 著者 | Schumacher, M.A. | | 登録日 | 2022-01-04 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7T1K
 
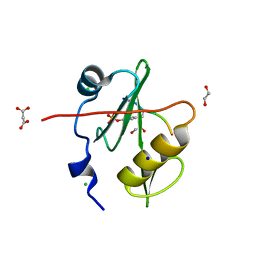 | | Crystal structure of a superbinder Fes SH2 domain (sFes1) in complex with a high affinity phosphopeptide | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, MALONATE ION, ... | | 著者 | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | 登録日 | 2021-12-02 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
7TDP
 
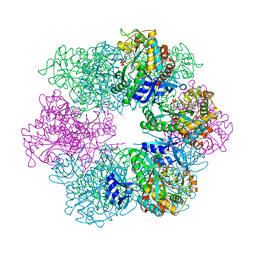 | |
7TF7
 
 | | S. aureus GS(12) - apo | | 分子名称: | Glutamine synthetase | | 著者 | Travis, B.A, Peck, J, Schumacher, M.A. | | 登録日 | 2022-01-06 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (2.13 Å) | | 主引用文献 | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TDV
 
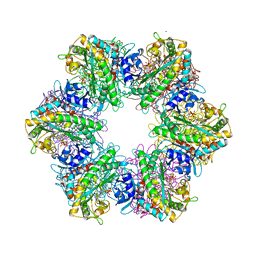 | |
7TEN
 
 | |
2J48
 
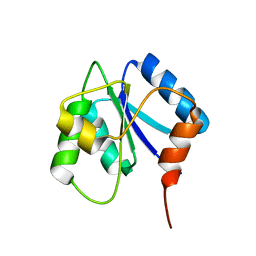 | |
4RUA
 
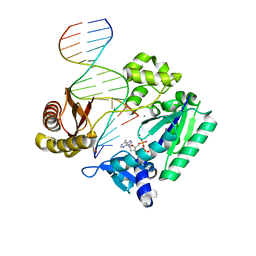 | | Crystal structure of Y-family DNA polymerase Dpo4 bypassing a MeFapy-dG adduct | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase IV, ... | | 著者 | Patra, A, Banerjee, S, Stone, M.P, Egli, M. | | 登録日 | 2014-11-18 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Structural Basis for Error-Free Bypass of the 5-N-Methylformamidopyrimidine-dG Lesion by Human DNA Polymerase eta and Sulfolobus solfataricus P2 Polymerase IV.
J.Am.Chem.Soc., 137, 2015
|
|
7TFD
 
 | | P. polymyxa GS(12) - apo | | 分子名称: | Glutamine synthetase, MAGNESIUM ION | | 著者 | Travis, B.A, Peck, J, Schumacher, M.A. | | 登録日 | 2022-01-06 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TGK
 
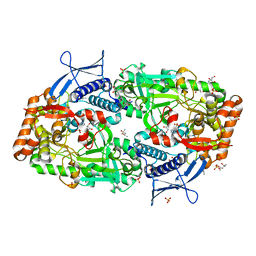 | | Crystal structure of ATP bound DesD, the desferrioxamine synthetase from the Streptomyces griseoflavus ferrimycin biosynthetic pathway | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Patel, K.D, Gulick, A.M. | | 登録日 | 2022-01-07 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | An acyl-adenylate mimic reveals the structural basis for substrate recognition by the iterative siderophore synthetase DesD.
J.Biol.Chem., 298, 2022
|
|
2ICP
 
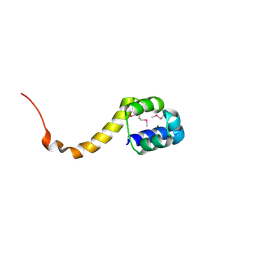 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 4.0. Northeast Structural Genomics Consortium TARGET ER390. | | 分子名称: | MAGNESIUM ION, antitoxin higa | | 著者 | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-09-13 | | 公開日 | 2006-09-26 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal structure of the bacterial antitoxin HigA from Escherichia coli.
To be Published
|
|
7SVB
 
 | | APE1 exonuclease substrate complex with 8oxoG opposite C | | 分子名称: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | 著者 | Whitaker, A.W, Freudenthal, B.D. | | 登録日 | 2021-11-18 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
4RX6
 
 | | Structure of B. subtilis GlnK-ATP complex to 2.6 Angstrom | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Nitrogen regulatory PII-like protein | | 著者 | Schumacher, M.A, Cuthbert, B, Tonthat, N, Chinnam, N.G, Whitfill, T. | | 登録日 | 2014-12-09 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5994 Å) | | 主引用文献 | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
4RZT
 
 | | Lac repressor engineered to bind sucralose, sucralose-bound tetramer | | 分子名称: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Lac repressor | | 著者 | Arbing, M.A, Cascio, D, Sawaya, M.R, Kosuri, S, Church, G.M. | | 登録日 | 2014-12-24 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Engineering an allosteric transcription factor to respond to new ligands.
Nat.Methods, 13, 2016
|
|
4RZS
 
 | | Lac repressor engineered to bind sucralose, unliganded tetramer | | 分子名称: | GLYCEROL, Lac repressor | | 著者 | Arbing, M.A, Cascio, D, Kosuri, S, Church, G.M. | | 登録日 | 2014-12-24 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Engineering an allosteric transcription factor to respond to new ligands.
Nat.Methods, 13, 2016
|
|
7T2T
 
 | |
4TLE
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | 分子名称: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | 著者 | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | 登録日 | 2014-05-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.936 Å) | | 主引用文献 | Atomic-scale origins of slowness in the cyanobacterial circadian clock
Science, 349, 2015
|
|
2VCE
 
 | | Characterization and engineering of the bifunctional N- and O- glucosyltransferase involved in xenobiotic metabolism in plants | | 分子名称: | 1,2-ETHANEDIOL, 2,4,5-trichlorophenol, HYDROQUINONE GLUCOSYLTRANSFERASE, ... | | 著者 | Brazier-Hicks, M, Offen, W.A, Gershater, M.C, Revett, T.J, Lim, E.K, Bowles, D.J, Davies, G.J, Edwards, R. | | 登録日 | 2007-09-20 | | 公開日 | 2007-10-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Characterization and Engineering of the Bifunctional N- and O-Glucosyltransferase Involved in Xenobiotic Metabolism in Plants.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1TFM
 
 | | CRYSTAL STRUCTURE OF A RIBOSOME INACTIVATING PROTEIN IN ITS NATURALLY INHIBITED FORM | | 分子名称: | 2-AMINO-4-ISOPROPYL-PTERIDINE-6-CARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Mishra, V, Bilgrami, S, Paramasivam, M, Yadav, S, Sharma, R.S, Kaur, P, Srinivasan, A, Babu, C.R, Singh, T.P. | | 登録日 | 2004-05-27 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF A RIBOSOME INACTIVATING PROTEIN IN ITS NATURALLY INHIBITED FORM
To be Published
|
|
2VL2
 
 | | Oxidized and reduced forms of human peroxiredoxin 5 | | 分子名称: | BENZOIC ACID, PEROXIREDOXIN-5 | | 著者 | Smeets, A, Declercq, J.P. | | 登録日 | 2008-01-08 | | 公開日 | 2008-08-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.925 Å) | | 主引用文献 | The Crystal Structures of Oxidized Forms of Human Peroxiredoxin 5 with an Intramolecular Disulfide Bond Confirm the Proposed Enzymatic Mechanism for Atypical 2-Cys Peroxiredoxins.
Arch.Biochem.Biophys., 477, 2008
|
|
8A4Y
 
 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with N-(2,3-dihydro-1H-inden-5-yl)acetamide | | 分子名称: | Host translation inhibitor nsp1, N-(2,3-dihydro-1H-inden-5-yl)acetamide | | 著者 | Borsatto, A, Galdadas, I, Ma, S, Damfo, S, Haider, S, Kozielski, F, Estarellas, C, Gervasio, F.L. | | 登録日 | 2022-06-13 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.099 Å) | | 主引用文献 | Revealing druggable cryptic pockets in the Nsp1 of SARS-CoV-2 and other beta-coronaviruses by simulations and crystallography.
Elife, 11, 2022
|
|
