7BTE
 
 | | Lifeact-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kumari, A, Ragunath, V.K, Sirajuddin, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into actin filament recognition by commonly used cellular actin markers.
Embo J., 39, 2020
|
|
6PUK
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-JYM72 | | Descriptor: | 1,2-dideoxy-1-{2,6-dioxo-5-[(1E)-3-oxobut-1-en-1-yl]-1,2,3,6-tetrahydropyrimidin-4-yl}-D-ribo-hexitol, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Awad, W, Keller, A.N, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
1TDG
 
 | | Complex of S130G SHV-1 beta-lactamase with tazobactam | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase SHV-1, ... | | Authors: | Sun, T, Bethel, C.R, Bonomo, R.A, Knox, J.R. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor-resistant class A beta-lactamases: consequences of the Ser130-to-Gly mutation seen in Apo and tazobactam structures of the SHV-1 variant
Biochemistry, 43, 2004
|
|
5KT2
 
 | | Teranry complex of human DNA polymerase iota(26-445) inserting dCMPNPP opposite template G in the presence of Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*TP*GP*GP*GP*GP*TP*CP*CP*T)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Choi, J.Y, Patra, A, Yeom, M, Lee, Y.S, Zhang, Q, Egli, M, Guengerich, F.P. | | Deposit date: | 2016-07-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase iota.
J.Biol.Chem., 291, 2016
|
|
6PV5
 
 | | Structure of CpGH84B | | Descriptor: | 1,2-ETHANEDIOL, Putative O-GlcNAcase nagJ | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural and functional analysis of four family 84 glycoside hydrolases from the opportunistic pathogen Clostridium perfringens.
Glycobiology, 30, 2019
|
|
1TEZ
 
 | | COMPLEX BETWEEN DNA AND THE DNA PHOTOLYASE FROM ANACYSTIS NIDULANS | | Descriptor: | 5'-D(*AP*TP*CP*GP*GP*CP*T*(TCP)P*CP*GP*C)-3', 5'-D(*TP*CP*GP*C)-3', 5'-D(P*CP*GP*AP*AP*GP*CP*CP*GP*A)-3', ... | | Authors: | Essen, L.-O, Carell, T, Mees, A, Klar, T. | | Deposit date: | 2004-05-26 | | Release date: | 2004-12-14 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair
Science, 306, 2004
|
|
2IU5
 
 | | Dihydroxyacetone kinase operon activator DhaS | | Descriptor: | HTH-TYPE DHAKLM OPERON TRANSCRIPTIONAL ACTIVATOR DHAS | | Authors: | Srinivas, A, Christen, S, Baumann, U, Erni, B. | | Deposit date: | 2006-05-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Regulation of the Dha Operon of Lactococcus Lactis: A Deviation from the Rule Followed by the Tetr Family of Transcription Regulators
J.Biol.Chem., 281, 2006
|
|
3ES2
 
 | |
3ZLS
 
 | | Crystal structure of MEK1 in complex with fragment 6 | | Descriptor: | 1H-PYRROLO[2,3-B]PYRIDINE-3-CARBOXYLIC ACID, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8A1G
 
 | | Structure of the SNX1-SNX5 complex | | Descriptor: | N-PROPANOL, Sorting nexin-1, Sorting nexin-5 | | Authors: | Lopez-Robles, C, Scaramuzza, S, Astorga-Simon, E.N, Banos-Mateos, S, Vidaurrazaga, A, Rojas, A.L, Castano, D, Hierro, A. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of the ESCPE-1 membrane coat.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6PSG
 
 | | Crystal Structure of Class D Beta-lactamase OXA-48 with Faropenem | | Descriptor: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, CHLORIDE ION, Class D Carbapenemase OXA-48, ... | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-07-12 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis for Substrate Specificity and Carbapenemase Activity of OXA-48 Class D beta-Lactamase.
Acs Infect Dis., 6, 2020
|
|
6PSW
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPo) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
8A1K
 
 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with ebsulfur analogue 15 | | Descriptor: | 1,2-ETHANEDIOL, 4,5-bis(chloranyl)-N-(2-hydroxyethyl)-2-sulfanyl-benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | de Munnik, M, Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-throughput screen with the l,d-transpeptidase Ldt Mt2 of Mycobacterium tuberculosis reveals novel classes of covalently reacting inhibitors.
Chem Sci, 14, 2023
|
|
2WG3
 
 | | Crystal structure of the complex between human hedgehog-interacting protein HIP and desert hedgehog without calcium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DESERT HEDGEHOG PROTEIN N-PRODUCT, ... | | Authors: | Bishop, B, Aricescu, A.R, Harlos, K, O'Callaghan, C.A, Jones, E.Y, Siebold, C. | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Hedgehog Ligand Sequestration by the Human Hedgehog-Interacting Protein Hip
Nat.Struct.Mol.Biol., 16, 2009
|
|
2J8T
 
 | | Human aldose reductase in complex with NADP and citrate at 0.82 angstrom | | Descriptor: | ALDO-KETO REDUCTASE FAMILY 1, MEMBER B1, CITRATE ANION, ... | | Authors: | Biadene, M, Hazemann, I, Cousido, A, Ginell, S, Sheldrick, G.M, Podjarny, A, Schneider, T.R. | | Deposit date: | 2006-10-27 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.82 Å) | | Cite: | The Atomic Resolution Structure of Human Aldose Reductase Reveals that Rearrangement of a Bound Ligand Allows the Opening of the Safety-Belt Loop.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
7T7F
 
 | | MA-1-206-OXA-23 25 minute complex | | Descriptor: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
1TDL
 
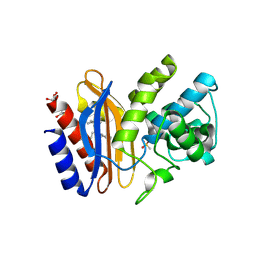 | | Structure of Ser130Gly SHV-1 beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Sun, T, Bethel, C.R, Bonomo, R.A, Knox, J.R. | | Deposit date: | 2004-05-23 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor-resistant class A beta-lactamases: consequences of the Ser130-to-Gly mutation seen in Apo and tazobactam structures of the SHV-1 variant
Biochemistry, 43, 2004
|
|
2VO8
 
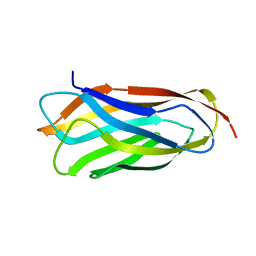 | | Cohesin module from Clostridium perfringens ATCC13124 family 33 glycoside hydrolase. | | Descriptor: | EXO-ALPHA-SIALIDASE | | Authors: | Gregg, K, Adams, J.J, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2008-02-08 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Clostridium Perfringens Toxin Complex Formation.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3ZLY
 
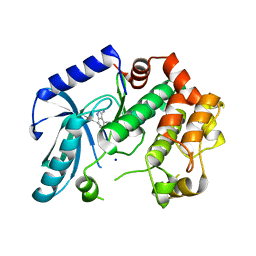 | | Crystal structure of MEK1 in complex with fragment 8 | | Descriptor: | 3-AMINO-1H-INDAZOLE-4-CARBONITRILE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3EVJ
 
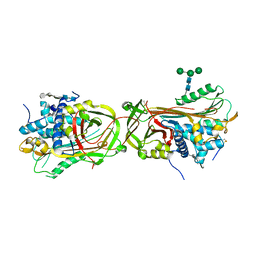 | | Intermediate structure of antithrombin bound to the natural pentasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, ... | | Authors: | Huntington, J.A, Belzar, K.J. | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The critical role of hinge-region expulsion in the induced-fit heparin binding mechanism of antithrombin.
J. Mol. Biol., 386, 2009
|
|
5KKC
 
 | | l-lactate dehydrogenase from rabbit muscle with the inhibitor 6DHNAD | | Descriptor: | L-lactate dehydrogenase A chain, SULFATE ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(5-aminocarbonyl-2~{H}-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Meneely, K.M, Moran, G.R, Lamb, A.L. | | Deposit date: | 2016-06-21 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Ligand binding phenomena that pertain to the metabolic function of renalase.
Arch.Biochem.Biophys., 612, 2016
|
|
2ID5
 
 | | Crystal Structure of the Lingo-1 Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine rich repeat neuronal 6A, ... | | Authors: | Mosyak, L, Wood, A, Dwyer, B, Johnson, M, Stahl, M.L, Somers, W.S. | | Deposit date: | 2006-09-14 | | Release date: | 2006-09-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition.
J.Biol.Chem., 281, 2006
|
|
3ENJ
 
 | | Structure of Pig Heart Citrate Synthase at 1.78 A resolution | | Descriptor: | CHLORIDE ION, CYSTEINE, Citrate synthase, ... | | Authors: | Larson, S.B, Day, J.S, Nguyen, C, Cudney, R, McPherson, A, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2008-09-25 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of pig heart citrate synthase at 1.78 A resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2IDR
 
 | | Crystal structure of translation initiation factor EIF4E from wheat | | Descriptor: | Eukaryotic translation initiation factor 4E-1 | | Authors: | Monzingo, A.F, Sadow, J, Dhaliwal, S, Lyon, A, Hoffman, D.W, Robertus, J.D, Browning, K.S. | | Deposit date: | 2006-09-15 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond.
Plant Physiol., 143, 2007
|
|
1TCO
 
 | | TERNARY COMPLEX OF A CALCINEURIN A FRAGMENT, CALCINEURIN B, FKBP12 AND THE IMMUNOSUPPRESSANT DRUG FK506 (TACROLIMUS) | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, FE (III) ION, ... | | Authors: | Griffith, J.P, Kim, J.L, Kim, E.E, Sintchak, M.D, Thomson, J.A, Fitzgibbon, M.J, Fleming, M.A, Caron, P.R, Hsiao, K, Navia, M.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex.
Cell(Cambridge,Mass.), 82, 1995
|
|
