1AEB
 
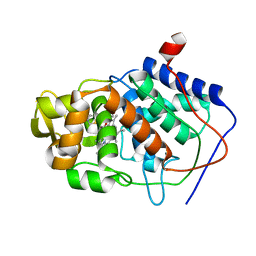 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (3-METHYLTHIAZOLE) | | Descriptor: | 3-METHYLTHIAZOLIUM ION, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
5KW4
 
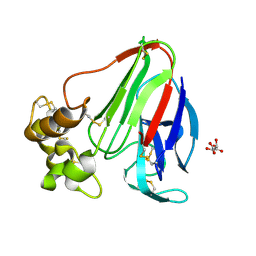 | | T. danielli thaumatin at 278K, Data set 2 | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S. | | Deposit date: | 2016-07-15 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
6GCZ
 
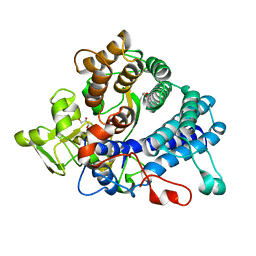 | | Laminarin binding SusD-like protein | | Descriptor: | GLYCEROL, Starch-binding associating with outer membrane | | Authors: | Mystowka, A, Robb, C.S, Hehemann, J.H. | | Deposit date: | 2018-04-20 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular recognition of the beta-glucans laminarin and pustulan by a SusD-like glycan-binding protein of a marine Bacteroidetes.
FEBS J., 285, 2018
|
|
1AEO
 
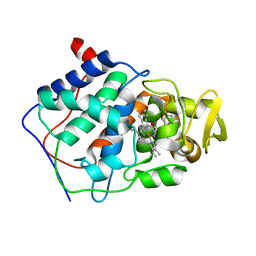 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (2-AMINOPYRIDINE) | | Descriptor: | 2-AMINOPYRIDINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Bunte, S.W, Rosenfeld, R, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
4UFM
 
 | | Mouse Galactocerebrosidase complexed with 1-deoxy-galacto-nojirimycin DGJ | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hill, C.H, Viuff, A.H, Spratley, S.J, Salamone, S, Christensen, S.H, Read, R.J, Moriarty, N.W, Jensen, H.H, Deane, J.E. | | Deposit date: | 2015-03-17 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Azasugar Inhibitors as Pharmacological Chaperones for Krabbe Disease.
Chem.Sci., 6, 2015
|
|
8SWJ
 
 | | Co-crystal structure of 53BP1 tandem Tudor domains in complex with UNC8531 | | Descriptor: | (3S)-N-(4'-carbamoyl[1,1'-biphenyl]-3-yl)-1-[4-(4-methylpiperazin-1-yl)pyridine-2-carbonyl]piperidine-3-carboxamide, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Shell, D.J, Foley, C, Pearce, K.H, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-crystal structure of 53BP1 tandem Tudor domains in complex with UNC8531
To be published
|
|
6W9P
 
 | | Tel26 Parallel Four-quartet G-quadruplex with K+ | | Descriptor: | DNA (5'-D(P*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*GP*T)-3'), POTASSIUM ION, SODIUM ION | | Authors: | McCarthy, S.E, Yatsunyk, L.A, Beseiso, D, Miao, J. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The first crystal structures of hybrid and parallel four-tetrad intramolecular G-quadruplexes.
Nucleic Acids Res., 50, 2022
|
|
5HYK
 
 | | Crystal structure of the complex PPARalpha/AL26-29 | | Descriptor: | 2-methyl-2-[4-(naphthalen-1-yl)phenoxy]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Lavecchia, A. | | Deposit date: | 2016-02-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for PPAR partial or full activation revealed by a novel ligand binding mode.
Sci Rep, 6, 2016
|
|
8ERF
 
 | |
8OX9
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2P active conformation with bound PC | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
8T5I
 
 | | Crystal structure of human WDR5 in complex with MR4397 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-[(2S)-1-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-1-oxopentan-2-yl]-3-[(1H-imidazol-1-yl)methyl]benzamide, ... | | Authors: | Kimani, S, Dong, A, Li, F, Loppnau, P, Ackloo, S, Vedadi, M, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-13 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human WDR5 in complex with MR4397
To be published
|
|
5KW8
 
 | | T. danielli thaumatin at 278K, Data set 5 | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S. | | Deposit date: | 2016-07-15 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
8P9O
 
 | | PCNA from Chaetomium thermophilum in complex with PolD3 peptide | | Descriptor: | Proliferating cell nuclear antigen, Synthetic peptide corresponding to amino acids 437 to 451 of PolD3 from Chaetomium thermophilum | | Authors: | Alphey, M.S, Wolford, C.B, MacNeill, S.A. | | Deposit date: | 2023-06-06 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Canonical binding of Chaetomium thermophilum DNA polymerase delta / zeta subunit PolD3 and flap endonuclease Fen1 to PCNA.
Front Mol Biosci, 10, 2023
|
|
7VML
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 1&2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
6GHG
 
 | | Variable heavy - variable light domain and Fab-arm CrossMabs with charged residue exchanges | | Descriptor: | Fab heavy chain, Fab light chain, HEXAETHYLENE GLYCOL, ... | | Authors: | Regula, J, Imhof-Jung, S, Molhoj, M, Benz, J, Ehler, A, Bujotzek, A, Schaefer, W, Klein, C. | | Deposit date: | 2018-05-07 | | Release date: | 2018-09-12 | | Last modified: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Variable heavy-variable light domain and Fab-arm CrossMabs with charged residue exchanges to enforce correct light chain assembly.
Protein Eng. Des. Sel., 31, 2018
|
|
1A8R
 
 | | GTP CYCLOHYDROLASE I (H112S MUTANT) IN COMPLEX WITH GTP | | Descriptor: | GTP CYCLOHYDROLASE I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Auerbach, G, Nar, H, Bracher, A, Bacher, A, Huber, R. | | Deposit date: | 1998-03-27 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biosynthesis of pteridines. Reaction mechanism of GTP cyclohydrolase I.
J.Mol.Biol., 326, 2003
|
|
3L08
 
 | | Structure of Pi3K gamma with a potent inhibitor: GSK2126458 | | Descriptor: | 2,4-difluoro-N-[2-methoxy-5-(4-pyridazin-4-ylquinolin-6-yl)pyridin-3-yl]benzenesulfonamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Elkins, P.A, Marrero, E.M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of GSK2126458, a Highly Potent Inhibitor of PI3K and the Mammalian Target of Rapamycin.
ACS Med Chem Lett, 1, 2010
|
|
5I08
 
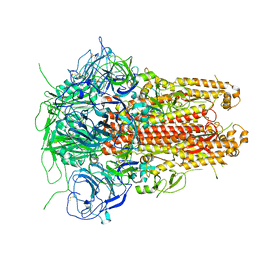 | | Prefusion structure of a human coronavirus spike protein | | Descriptor: | Spike glycoprotein,Foldon chimera | | Authors: | Kirchdoerfer, R.N, Cottrell, C.A, Wang, N, Pallesen, J, Yassine, H.M, Turner, H.L, Corbett, K.S, Graham, B.S, McLellan, J.S, Ward, A.B. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pre-fusion structure of a human coronavirus spike protein.
Nature, 531, 2016
|
|
1ACY
 
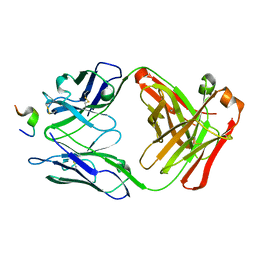 | |
5ZJ9
 
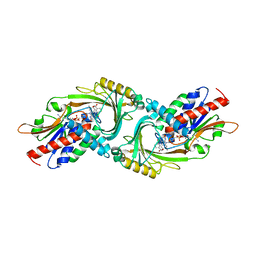 | | human D-amino acid oxidase complexed with 5-chlorothiophene-3-carboxylic acid | | Descriptor: | 5-chloro thiophene-3-carboxylic acid, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kato, Y, Hin, N, Maita, N, Thomas, A.G, Kurosawa, S, Rojas, C, Yorita, K, Slusher, B.S, Fukui, K, Tsukamoto, T. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for potent inhibition of d-amino acid oxidase by thiophene carboxylic acids
Eur J Med Chem, 159, 2018
|
|
1AD8
 
 | | COMPLEX OF THROMBIN WITH AND INHIBITOR CONTAINING A NOVEL P1 MOIETY | | Descriptor: | HIRUDIN (53-65) PEPTIDE, SODIUM ION, THROMBIN (LARGE SUBUNIT), ... | | Authors: | Schreuder, H, Tardif, C, Malikayil, J.A. | | Deposit date: | 1997-02-24 | | Release date: | 1997-11-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular design and characterization of an alpha-thrombin inhibitor containing a novel P1 moiety.
Biochemistry, 36, 1997
|
|
6QJP
 
 | | Cryo-EM structure of heparin-induced 2N4R tau jagged filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
7ZB6
 
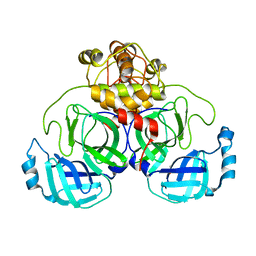 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) variant C44S at 2.12 A resolution | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Paknia, E, Rabe von Pappenheim, F, Funk, L.-M, Tittmann, K, Chari, A. | | Deposit date: | 2022-03-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Multiple redox switches of the SARS-CoV-2 main protease in vitro provide opportunities for drug design.
Nat Commun, 15, 2024
|
|
6QCG
 
 | |
7SJ3
 
 | | Structure of CDK4-Cyclin D3 bound to abemaciclib | | Descriptor: | Cyclin-dependent kinase 4, G1/S-specific cyclin-D3, N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py rimidin-2-amine | | Authors: | Hilgers, M.T, Pelletier, L.A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of CDK4-Cyclin D3 bound to abemaciclib
To Be Published
|
|
