5AYF
 
 | | Crystal structure of SET7/9 in complex with cyproheptadine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(dibenzo[1,2-a:2',1'-d][7]annulen-11-ylidene)-1-methyl-piperidine, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Niwa, H, Handa, N, Takemoto, Y, Ito, A, Tomabechi, Y, Umehara, T, Shirouzu, M, Yoshida, M, Yokoyama, S. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Identification of Cyproheptadine as an Inhibitor of SET Domain Containing Lysine Methyltransferase 7/9 (Set7/9) That Regulates Estrogen-Dependent Transcription
J.Med.Chem., 59, 2016
|
|
1U4S
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with 2,6-naphthalenedisulphonic acid | | Descriptor: | L-lactate dehydrogenase, NAPHTHALENE-2,6-DISULFONIC ACID | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
1U62
 
 | |
3ZLV
 
 | | Crystal structure of mouse acetylcholinesterase in complex with tabun and HI-6 | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Artursson, E, Andersson, P.O, Akfur, C, Linusson, A, Borjegren, S, Ekstrom, F. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic-Site Conformational Equilibrium in Nerve-Agent Adducts of Acetylcholinesterase; Possible Implications for the Hi-6 Antidote Substrate Specificity.
Biochem.Pharmacol., 85, 2013
|
|
6ERY
 
 | |
2P5Y
 
 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell, J.T, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
6E97
 
 | |
2P02
 
 | | Crystal structure of the alpha subunit of human S-adenosylmethionine synthetase 2 | | Descriptor: | CHLORIDE ION, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthetase isoform type-2 | | Authors: | Papagrigoriou, E, Shafqat, N, Rojkova, A, Niessen, F.H, Kavanagh, K.L, von Delft, F, Gorrec, F, Ugochukwu, E, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal structure of the alpha subunit of human S-adenosylmethionine synthetase 2
To be Published
|
|
3ZID
 
 | |
8PW8
 
 | | Crystal structure of the human METTL3-METTL14 in complex with a bisubstrate analogue (BA2) | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[2-[[9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-6-yl]amino]ethyl]amino]-2-azanyl-butanoic acid, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Etheve-Quelquejeu, M, Caflisch, A. | | Deposit date: | 2023-07-19 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The catalytic mechanism of the RNA methyltransferase METTL3.
Elife, 12, 2024
|
|
8POE
 
 | | Structure of tissue-specific lipid scramblase ATG9B homotrimer, refined with C3 symmetry applied | | Descriptor: | Autophagy-related protein 9B | | Authors: | Chiduza, G.N, Pye, V.E, Tooze, S.A, Cherepanov, P. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | ATG9B is a tissue-specific homotrimeric lipid scramblase that can compensate for ATG9A.
Autophagy, 20, 2024
|
|
5B18
 
 | | Crystal Structure of a Darunavir Resistant HIV-1 Protease | | Descriptor: | ACETATE ION, CHLORIDE ION, Protease | | Authors: | Suzuki, K, Ode, H, Nakashima, M, Sugiura, W, Watanabe, N, Suzuki, A, Iwatani, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unique Flap Conformation in an HIV-1 Protease with High-Level Darunavir Resistance
Front Microbiol, 7, 2016
|
|
5B7V
 
 | | Human FGFR1 kinase in complex with CH5183284 | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, [5-amino-1-(2-methyl-1H-benzimidazol-6-yl)-1H-pyrazol-4-yl](1H-indol-2-yl)methanone | | Authors: | Fukami, T.A, Lukacs, C.M, Janson, C. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor
Mol.Cancer Ther., 13, 2014
|
|
8PWA
 
 | | Crystal structure of the human METTL3-METTL14 in complex with a bisubstrate analogue (BA4) | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[[9-[(2~{R},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-7~{H}-purin-6-yl]amino]propyl]amino]-2-azanyl-butanoic acid, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Etheve-Quelquejeu, M, Caflisch, A. | | Deposit date: | 2023-07-19 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The catalytic mechanism of the RNA methyltransferase METTL3.
Elife, 12, 2024
|
|
8PWB
 
 | | Crystal structure of the human METTL3-METTL14 in complex with a bisubstrate analogue (BA6) | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(7~{H}-purin-6-ylcarbamoyl)amino]-2-azanyl-butanoic acid, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Etheve-Quelquejeu, M, Caflisch, A. | | Deposit date: | 2023-07-19 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic mechanism of the RNA methyltransferase METTL3.
Elife, 12, 2024
|
|
4MIN
 
 | |
4MVN
 
 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | Descriptor: | Serine protease splA, [(1S)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Zdzalik, M, Burchacka, E, Niemczyk, J.S, Pustelny, K, Popowicz, G.M, Wladyka, B, Dubin, A, Potempa, J, Sienczyk, M, Dubin, G, Oleksyszyn, J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
8PW9
 
 | | Crystal structure of the human METTL3-METTL14 in complex with a bisubstrate analogue (BA1) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[2-[[9-[(2~{R},3~{R},4~{S},5~{S})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-6-yl]amino]ethylamino]methyl]oxolane-3,4-diol, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Etheve-Quelquejeu, M, Caflisch, A. | | Deposit date: | 2023-07-19 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The catalytic mechanism of the RNA methyltransferase METTL3.
Elife, 12, 2024
|
|
3ZL1
 
 | | A thiazolyl-mannoside bound to FimH, monoclinic space group | | Descriptor: | CHLORIDE ION, N-{5-[(1R)-1-hydroxyethyl]-1,3-thiazol-2-yl}-alpha-D-mannopyranosylamine, PROTEIN FIMH | | Authors: | Brument, S, Sivignon, A, Dumych, T.I, Moreau, N, Roos, G, Guerardel, Y, Chalopin, T, Deniaud, D, Bilyy, R.O, Darfeuille-Michaud, A, Bouckaert, J, Gouin, S.G. | | Deposit date: | 2013-01-27 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Thiazolylaminomannosides as Potent Antiadhesives of Type 1 Piliated Escherichia Coli Isolated from Crohn'S Disease Patients.
J.Med.Chem., 56, 2013
|
|
6EGK
 
 | | T181N Cucumene Synthase | | Descriptor: | Cucumene Synthase | | Authors: | Blank, P.N, Pemberton, T.A, Christianson, D.W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Cucumene Synthase, a Terpenoid Cyclase That Generates a Linear Triquinane Sesquiterpene.
Biochemistry, 57, 2018
|
|
3ZOA
 
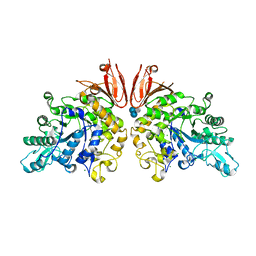 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
6EH0
 
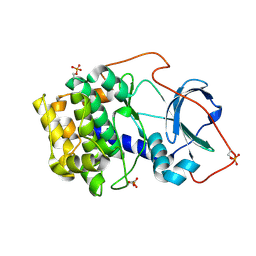 | |
1UCG
 
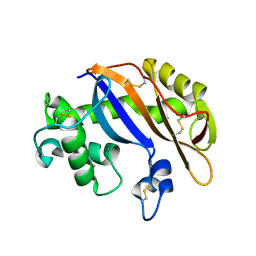 | | Crystal structure of Ribonuclease MC1 N71T mutant | | Descriptor: | MANGANESE (II) ION, Ribonuclease MC | | Authors: | Suzuki, A, Numata, T, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2003-04-14 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the ribonuclease MC1 mutants N71T and N71S in complex with 5'-GMP: structural basis for alterations in substrate specificity
Biochemistry, 42, 2003
|
|
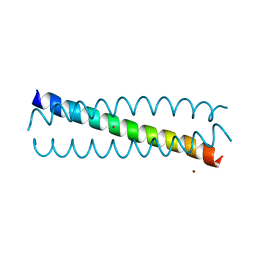
6EGP
 
 | |
8Q1B
 
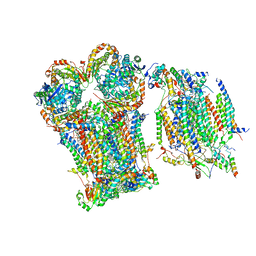 | | III2-IV1 respiratory supercomplex from S. pombe | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Moe, A, Brzezinski, P. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of the S. pombe III-IV-cyt c supercomplex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
