5UFI
 
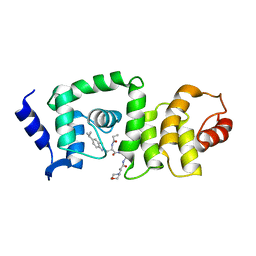 | | DCN1 bound to DI-591 | | Descriptor: | DCN1-like protein 1, N-[(1S)-1-cyclohexyl-2-{[3-(morpholin-4-yl)propanoyl]amino}ethyl]-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J. | | Deposit date: | 2017-01-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A potent small-molecule inhibitor of the DCN1-UBC12 interaction that selectively blocks cullin 3 neddylation.
Nat Commun, 8, 2017
|
|
4I80
 
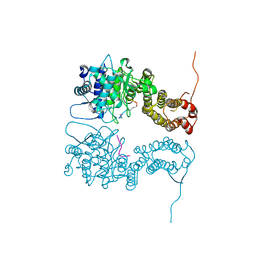 | |
3SME
 
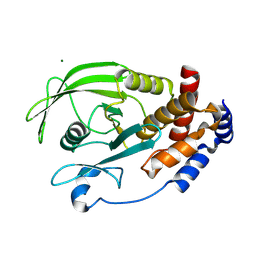 | | Structure of PTP1B inactivated by H2O2/bicarbonate | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Tanner, J.J, Singh, H. | | Deposit date: | 2011-06-27 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Biological Buffer Bicarbonate/CO(2) Potentiates H(2)O(2)-Mediated Inactivation of Protein Tyrosine Phosphatases.
J.Am.Chem.Soc., 133, 2011
|
|
6B5Q
 
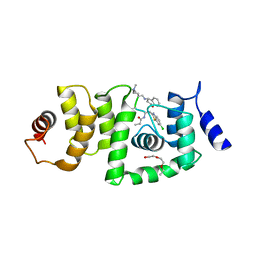 | | DCN1 bound to 38 | | Descriptor: | DCN1-like protein 1, Peptidomimetic Inhibitors DI-591, TRIETHYLENE GLYCOL | | Authors: | Stuckey, J. | | Deposit date: | 2017-09-29 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | High-Affinity Peptidomimetic Inhibitors of the DCN1-UBC12 Protein-Protein Interaction.
J. Med. Chem., 61, 2018
|
|
2Q6J
 
 | | Crystal Structure of Estrogen Receptor alpha Complexed to a B-N Substituted Ligand | | Descriptor: | 4-[(DIMESITYLBORYL)(2,2,2-TRIFLUOROETHYL)AMINO]PHENOL, Estrogen receptor, GRIP peptide | | Authors: | Zhou, H, Nettles, K.W, Bruning, J.B, Kim, Y, Joachimiak, A, Sharma, S, Carlson, K.E, Stossi, F, Katzenellenbogen, B.S, Greene, G.L, Katzenellenbogen, J.A. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Elemental isomerism: a boron-nitrogen surrogate for a carbon-carbon double bond increases the chemical diversity of estrogen receptor ligands
Chem.Biol., 14, 2007
|
|
3LFL
 
 | | Crystal Structure of human Glutathione Transferase Omega 1, delta 155 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLUTATHIONE, Glutathione S-transferase omega-1 | | Authors: | Brock, J. | | Deposit date: | 2010-01-18 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel folding and stability defects cause a deficiency of human glutathione transferase omega 1.
J.Biol.Chem., 286, 2011
|
|
3OSV
 
 | | The crytsal structure of FLGD from P. Aeruginosa | | Descriptor: | Flagellar basal-body rod modification protein FlgD, GLYCEROL | | Authors: | Wang, D, Luo, M, Niu, S. | | Deposit date: | 2010-09-10 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a novel dimer form of FlgD from P. aeruginosa PAO1
Proteins, 79, 2011
|
|
3SP7
 
 | | Crystal Structure of Bcl-xL bound to BM903 | | Descriptor: | 5-(4-chlorophenyl)-4-{3-[4-(4-{[(4-{[(2R)-4-(dimethylamino)-1-(phenylsulfanyl)butan-2-yl]amino}-3-nitrophenyl)sulfonyl]amino}phenyl)piperazin-1-yl]phenyl}-1,2-dimethyl-1H-pyrrole-3-carboxylic acid, ACETATE ION, Bcl-2-like protein 1, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based design of a new class of potent Bcl-2/Bcl-xL inhibitors
To be Published
|
|
3SPF
 
 | | Crystal Structure of Bcl-xL bound to BM501 | | Descriptor: | 4-(4-chlorophenyl)-1-[(3S)-3,4-dihydroxybutyl]-N-[3-(4-methylpiperazin-1-yl)propyl]-3-phenyl-1H-pyrrole-2-carboxamide, Bcl-2-like protein 1, GLYCEROL | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2011-07-01 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of Bcl-2 and Bcl-xL Inhibitors with Subnanomolar Binding Affinities Based upon a New Scaffold.
J.Med.Chem., 55, 2012
|
|
3VLN
 
 | | Human Glutathione Transferase O1-1 C32S Mutant in Complex with Ascorbic Acid | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ASCORBIC ACID, ... | | Authors: | Brock, J, Board, P.G, Oakley, A.J. | | Deposit date: | 2011-12-02 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the dehydroascorbate reductase activity of human omega-class glutathione transferases.
J.Mol.Biol., 420, 2012
|
|
4XEO
 
 | |
6FWB
 
 | | Crystal structure of Mat2A at 1.79 Angstron resolution | | Descriptor: | GLYCEROL, S-adenosylmethionine synthase isoform type-2, SODIUM ION, ... | | Authors: | Zhou, A, Wei, Z, Bai, J, Wang, H. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-27 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of a natural inhibitor of methionine adenosyltransferase 2A regulating one-carbon metabolism in keratinocytes.
Ebiomedicine, 39, 2019
|
|
8TYP
 
 | | Complement Protease C1s Inhibited by 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide | | Descriptor: | 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide, Complement C1s subcomponent, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of the C1s Protease and the Classical Complement Pathway by 6-(4-Phenylpiperazin-1-yl)Pyridine-3-Carboximidamide and Chemical Analogs.
J Immunol., 212, 2024
|
|
7RRB
 
 | | IDO1 IN COMPLEX WITH COMPOUND 9 | | Descriptor: | 3-[4-(6-cyclopropylpyridin-3-yl)phenyl]-N-(4-fluorophenyl)oxetane-3-carboxamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-16 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Oxetane Promise Delivered: Discovery of Long-Acting IDO1 Inhibitors Suitable for Q3W Oral or Parenteral Dosing.
J.Med.Chem., 65, 2022
|
|
7RRC
 
 | | IDO1 IN COMPLEX WITH COMPOUND 14 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(4-fluorophenyl)-3-{4-[4-(hydroxymethyl)-6-(trifluoromethyl)pyridin-3-yl]phenyl}oxetane-3-carboxamide | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-16 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Oxetane Promise Delivered: Discovery of Long-Acting IDO1 Inhibitors Suitable for Q3W Oral or Parenteral Dosing.
J.Med.Chem., 65, 2022
|
|
6X5Y
 
 | | IDO1 in complex with compound 4 | | Descriptor: | 4-fluoro-N-{1-[5-(2-methylpyrimidin-4-yl)-5,6,7,8-tetrahydro-1,5-naphthyridin-2-yl]cyclopropyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-05-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Utilization of MetID and Structural Data to Guide Placement of Spiro and Fused Cyclopropyl Groups for the Synthesis of Low Dose IDO1 Inhibitors
To Be Published
|
|
7RRD
 
 | | IDO1 IN COMPLEX WITH COMPOUND S-1 | | Descriptor: | 3-[4-(1H-benzimidazol-2-yl)phenyl]-N-(4-fluorophenyl)oxetane-3-carboxamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Oxetane promise delivered: discovery of long acting IDO1 inhibitors suitable for Q3W oral or parenteral dosing
To Be Published
|
|
6SZE
 
 | |
6SZJ
 
 | |
6PU7
 
 | | Human IDO1 in complex with compound 17 (N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide) | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-07-17 | | Release date: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Amino-cyclobutarene-derived Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitors for Cancer Immunotherapy.
Acs Med.Chem.Lett., 10, 2019
|
|
6CW7
 
 | | E. coli DHFR product complex with (6S)-5,6,7,8-TETRAHYDROFOLATE | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-03-30 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
6CYV
 
 | | E. coli DHFR ternary complex with NADP and dihydrofolate | | Descriptor: | DIHYDROFOLIC ACID, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-04-06 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
6CXK
 
 | | E. coli DHFR substrate complex with Dihydrofolate | | Descriptor: | CHLORIDE ION, DIHYDROFOLIC ACID, Dihydrofolate reductase, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-04-03 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.108 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
4U97
 
 | | Crystal Structure of Asymmetric IRAK4 Dimer | | Descriptor: | Interleukin-1 receptor-associated kinase 4, STAUROSPORINE, SULFATE ION | | Authors: | Ferrao, R, Wu, H. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | IRAK4 Dimerization and trans-Autophosphorylation Are Induced by Myddosome Assembly.
Mol.Cell, 55, 2014
|
|
4U9A
 
 | |
