6XC4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 heavy chain, CC12.3 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC3
 
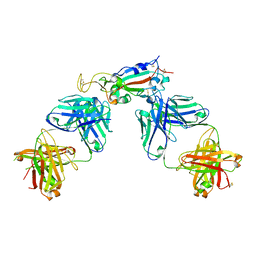 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.1 and CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.3 and CR3022 | | Descriptor: | CC12.3 heavy chain, CC12.3 light chain, CR3022 heavy chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC2
 
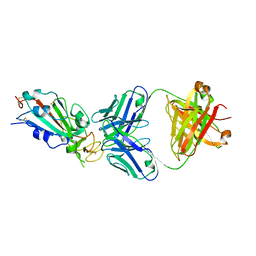 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.112 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6C3N
 
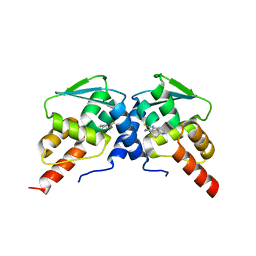 | | Crystal structure of BCL6 BTB domain in complex with compound 7CC5 | | Descriptor: | B-cell lymphoma 6 protein, N-(2-phenylethyl)-N'-pyridin-3-ylthiourea | | Authors: | Linhares, B, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53170586 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
6CQ1
 
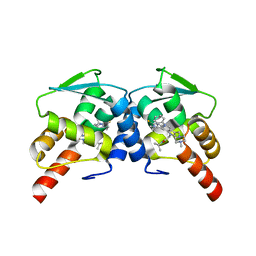 | | BCL6 BTB domain in complex with 15a | | Descriptor: | 2-{6-({[2-(1H-indol-3-yl)ethyl]carbamothioyl}amino)-3-[(4-methylpiperazin-1-yl)methyl]-1H-indol-1-yl}-N-(propan-2-yl)acetamide, B-cell lymphoma 6 protein | | Authors: | Linhares, B.M, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69921041 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
6DCV
 
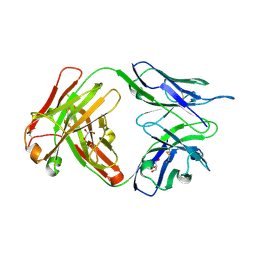 | | Crystal structure of human anti-tau antibody CBTAU-27.1 | | Descriptor: | GLYCEROL, Light chain of CBTAU27.1 Fab, heavy chain of CBTAU-27.1 Fab | | Authors: | Zhu, X, Zhang, H, Wilson, I.A. | | Deposit date: | 2018-05-08 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
6DCW
 
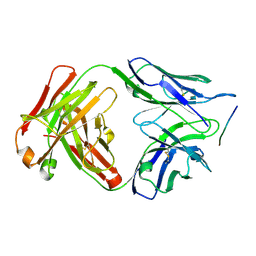 | |
5CP7
 
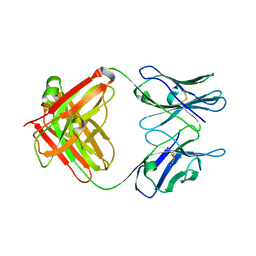 | | Crystal Structure of an Antigen-Binding Fragment of Monoclonal Antibody against Sulfonamides | | Descriptor: | Heavy Chain of Antigen-Binding Fragment of Monoclonal Antibody of 4C7, Light Chain of Antigen-Binding Fragment of Monoclonal Antibody of 4C7 | | Authors: | Wang, Z, Shen, J, Li, C, Li, Y, Wen, K, Yu, X, Zhang, X. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Class-specific Monoclonal Antibodies and Dihydropteroate Synthase in Bioassays used for the Detection of Sulfonamides: Structural Insights into Recognition Diversity.
Anal. Chem., 91, 2019
|
|
5CP3
 
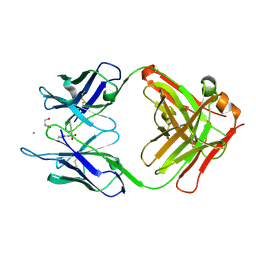 | | Crystal Structure of an Antigen-Binding Fragment of Monoclonal Antibody against Sulfonamides in Complex with Sulfathiazole | | Descriptor: | 4-amino-N-(1,3-thiazol-2-yl)benzenesulfonamide, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Z, Shen, J, Li, C, Li, Y, Wen, K, Yu, X, Zhang, X. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Class-specific Monoclonal Antibodies and Dihydropteroate Synthase in Bioassays used for the Detection of Sulfonamides: Structural Insights into Recognition Diversity.
Anal. Chem., 91, 2019
|
|
5CJQ
 
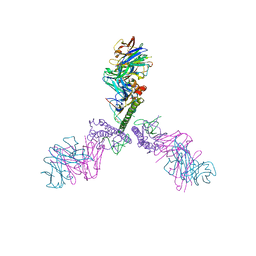 | |
5CJS
 
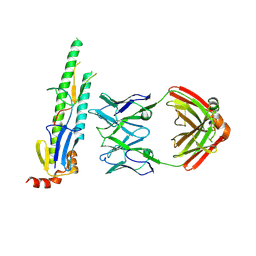 | |
8I3U
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 14B1, Light chain of Fab 14B1, ... | | Authors: | Li, Z, Yu, F, Cao, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
8I3S
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain od Fab 7B3, Light chain of Fab 7B3, ... | | Authors: | Li, Z, Yu, F, Cao, S, ZHao, H. | | Deposit date: | 2023-01-17 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
8IW0
 
 | | Crystal structure of the KANK1/liprin-beta1 complex | | Descriptor: | Liprin-beta-1,KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Zhang, J, Chen, S, Wei, Z, Yu, C. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
7KBH
 
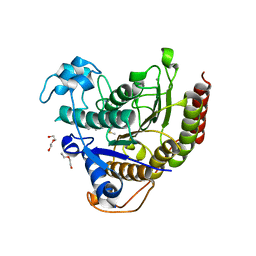 | |
7KBG
 
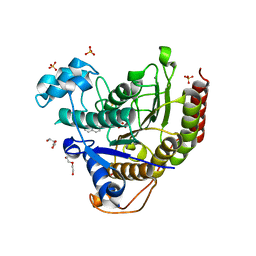 | |
6U02
 
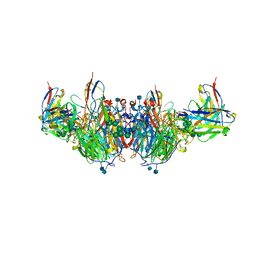 | | CryoEM-derived model of NA-63 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab-63 Heavy Chain, Fab-63 Light Chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6UWI
 
 | | Crystal structure of the Clostridium difficile translocase CDTb | | Descriptor: | ADP-ribosyltransferase binding component, CALCIUM ION | | Authors: | Pozharski, E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the cell-binding component of theClostridium difficilebinary toxin reveals a di-heptamer macromolecular assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UWT
 
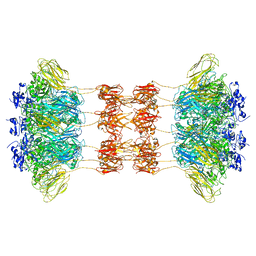 | |
6UWR
 
 | |
6UWO
 
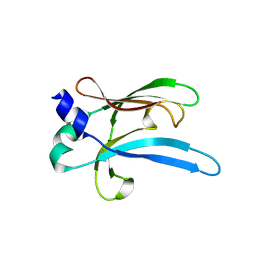 | |
6V52
 
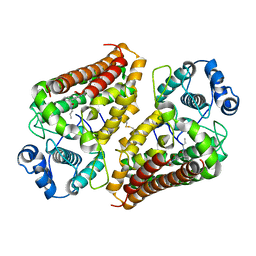 | | IDO1 IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 3-chloro-N-{4-[1-(propylcarbamoyl)cyclobutyl]phenyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Koenig, K.V, Augustin, M.A. | | Deposit date: | 2019-12-03 | | Release date: | 2020-04-08 | | Last modified: | 2020-04-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Strategic Incorporation of Polarity in Heme-Displacing Inhibitors of Indoleamine-2,3-dioxygenase-1 (IDO1).
Acs Med.Chem.Lett., 11, 2020
|
|
8IVZ
 
 | | Crystal structure of talin R7 in complex with KANK1 KN motif | | Descriptor: | KN motif and ankyrin repeat domains 1, Talin-1 | | Authors: | Xu, Y, Li, K, Wei, Z, Cong, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
8IW5
 
 | | Crystal structure of liprin-beta H2H3 dimer | | Descriptor: | CALCIUM ION, Liprin-beta-1 | | Authors: | Zhang, J, Chen, S, Wei, Z. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
