3RRU
 
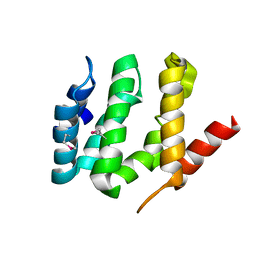 | | X-ray crystal structure of the VHS domain of human TOM1-like protein, Northeast Structural Genomics Consortium Target HR3050E | | Descriptor: | TOM1L1 protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, H, Ciccosanti, C, Patel, D, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-30 | | Release date: | 2011-06-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of the VHS domain of human TOM1-like protein, Northeast Structural Genomics Consortium Target HR3050E
To be Published
|
|
3RD4
 
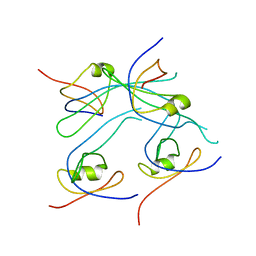 | | Crystal structure of PROPEN_03304 from Proteus penneri ATCC 35198 Northeast Structural Genomics Consortium target id PvR55 | | Descriptor: | uncharacterized protein | | Authors: | Seetharaman, J, Min, S, Kuzin, A, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of PROPEN_03304 from Proteus penneri ATCC 35198 Northeast Structural Genomics Consortium target id PvR55
To be Published
|
|
1NDF
 
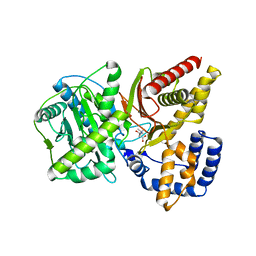 | | Carnitine Acetyltransferase in Complex with Carnitine | | Descriptor: | CARNITINE, Carnitine Acetyltransferase | | Authors: | Jogl, G, Tong, L. | | Deposit date: | 2002-12-09 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Carnitine Acetyltransferase and Implications for the Catalytic Mechanism and Fatty Acid Transport
Cell(Cambridge,Mass.), 112, 2003
|
|
1NJU
 
 | | Complex structure of HCMV Protease and a peptidomimetic inhibitor | | Descriptor: | Assemblin, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | Authors: | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
1NJT
 
 | | COMPLEX STRUCTURE OF HCMV PROTEASE AND A PEPTIDOMIMETIC INHIBITOR | | Descriptor: | CHLORIDE ION, Capsid protein P40, Peptidomimetic Inhibitor | | Authors: | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
1NF0
 
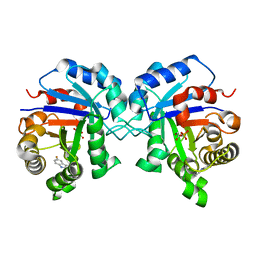 | | Triosephosphate Isomerase in Complex with DHAP | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, triosephosphate isomerase | | Authors: | Jogl, G, Rozovsky, S, McDermott, A.E, Tong, L. | | Deposit date: | 2002-12-12 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimal alignment for enzymatic proton transfer: Structure of the
Michaelis complex of triosephosphate isomerase at 1.2-A resolution
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3RUP
 
 | |
3RV4
 
 | |
3RV3
 
 | |
1NDI
 
 | | Carnitine Acetyltransferase in complex with CoA | | Descriptor: | COENZYME A, Carnitine Acetyltransferase | | Authors: | Jogl, G, Tong, L. | | Deposit date: | 2002-12-09 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Carnitine Acetyltransferase and Implications for the Catalytic Mechanism and Fatty Acid Transport
Cell(Cambridge,Mass.), 112, 2003
|
|
1NKM
 
 | | Complex structure of HCMV Protease and a peptidomimetic inhibitor | | Descriptor: | Assemblin, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | Authors: | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | Deposit date: | 2003-01-03 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
1NKK
 
 | | COMPLEX STRUCTURE OF HCMV PROTEASE AND A PEPTIDOMIMETIC INHIBITOR | | Descriptor: | Capsid protein P40, Peptidomimetic inhibitor | | Authors: | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | Deposit date: | 2003-01-03 | | Release date: | 2003-02-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
1NEY
 
 | | Triosephosphate Isomerase in Complex with DHAP | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, triosephosphate isomerase | | Authors: | Jogl, G, Rozovsky, S, McDermott, A.E, Tong, L. | | Deposit date: | 2002-12-12 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Optimal alignment for enzymatic proton transfer: Structure of the
Michaelis complex of triosephosphate isomerase at 1.2-A resolution.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NDB
 
 | | Crystal structure of Carnitine Acetyltransferase | | Descriptor: | Carnitine Acetyltransferase | | Authors: | Jogl, G, Tong, L. | | Deposit date: | 2002-12-09 | | Release date: | 2003-01-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Carnitine Acetyltransferase and Implications for the Catalytic Mechanism and Fatty Acid Transport
Cell(Cambridge,Mass.), 112, 2003
|
|
3SM3
 
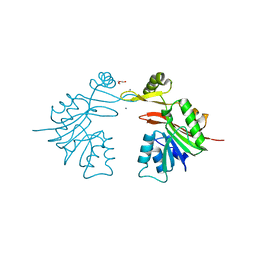 | | Crystal Structure of SAM-dependent methyltransferases Q8PUK2_METMA from Methanosarcina mazei. Northeast Structural Genomics Consortium Target MaR262. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, SAM-dependent methyltransferases | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Snyder, A, Patel, P, Xiao, R, Ciccosanti, C, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Crystal Structure of SAM-dependent methyltransferases Q8PUK2_METMA from Methanosarcina mazei.
To be Published, 2009
|
|
1OD2
 
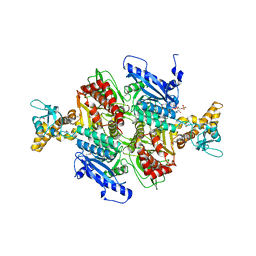 | | Acetyl-CoA Carboxylase Carboxyltransferase Domain | | Descriptor: | ACETYL COENZYME *A, ACETYL-COENZYME A CARBOXYLASE, ADENINE | | Authors: | Zhang, H, Yang, Z, Shen, Y, Tong, L. | | Deposit date: | 2003-02-12 | | Release date: | 2003-04-03 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase.
Science, 299, 2003
|
|
1NXH
 
 | | X-RAY STRUCTURE: NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT87 | | Descriptor: | MTH396 protein | | Authors: | Khayat, R, Savchenko, A, Edwards, A, Arowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-10 | | Release date: | 2004-02-24 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-RAY STRUCTURE OF MTH396
To be Published
|
|
3BEY
 
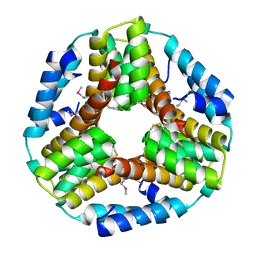 | | Crystal structure of the protein O27018 from Methanobacterium thermoautotrophicum. Northeast Structural Genomics Consortium target TT217 | | Descriptor: | Conserved protein O27018 | | Authors: | Kuzin, A.P, Gu, J, Xu, X, Neely, H, Forouhar, F, Owens, L, Mao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-18 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the protein O27018 from Methanobacterium thermoautotrophicum.
To be Published
|
|
1NXZ
 
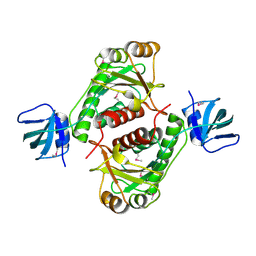 | | X-Ray Crystal Structure of Protein yggj_haein of Haemophilus influenzae. Northeast Structural Genomics Consortium Target IR73. | | Descriptor: | Hypothetical protein HI0303 | | Authors: | Forouhar, F, Shen, J, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional assignment based on structural analysis: crystal structure of the yggJ protein (HI0303) of Haemophilus influenzae reveals an RNA methyltransferase with a deep trefoil knot.
Proteins, 53, 2003
|
|
1NXU
 
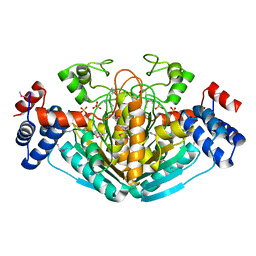 | | CRYSTAL STRUCTURE OF E. COLI HYPOTHETICAL OXIDOREDUCTASE YIAK NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82. | | Descriptor: | Hypothetical oxidoreductase yiaK, SULFATE ION | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Shastry, R, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
1OD4
 
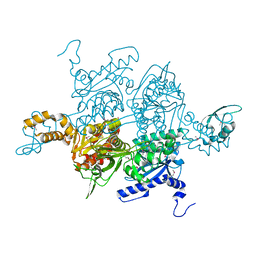 | | Acetyl-CoA Carboxylase Carboxyltransferase Domain | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE, ADENINE | | Authors: | Zhang, H, Yang, Z, Shen, Y, Tong, L. | | Deposit date: | 2003-02-12 | | Release date: | 2003-04-03 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase.
Science, 299, 2003
|
|
1O77
 
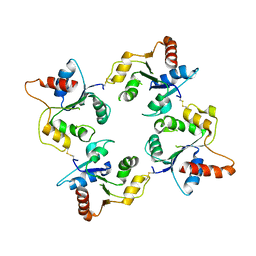 | | CRYSTAL STRUCTURE OF THE C713S MUTANT OF THE TIR DOMAIN OF HUMAN TLR2 | | Descriptor: | TOLL-LIKE RECEPTOR 2 | | Authors: | Tao, X, Xu, Y, Ye, Z, Beg, A.A, Tong, L. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | An Extensively Associated Dimer in the Structure of the C713S Mutant of the Tir Domain of Human Tlr2
Biochem.Biophys.Res.Commun., 299, 2002
|
|
3SYX
 
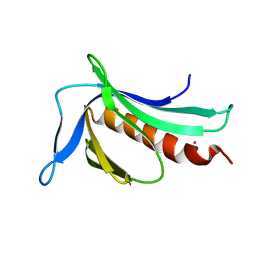 | | Crystal Structure of the WH1 domain from human sprouty-related, EVH1 domain-containing protein. Northeast Structural Genomics Consortium Target HR5538B. | | Descriptor: | Sprouty-related, EVH1 domain-containing protein 1, YTTRIUM (III) ION | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Crystal Structure of the WH1 domain from human sprouty-related, EVH1 domain-containing protein.
To be Published
|
|
2GWG
 
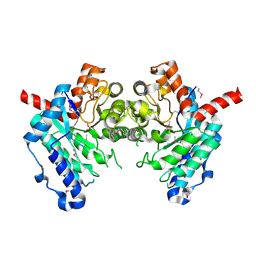 | | Crystal Structure of 4-Oxalomesaconate Hydratase, LigJ, from Rhodopseudomonas palustris, Northeast Structural Genomics Target RpR66. | | Descriptor: | 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Forouhar, F, Abashidze, M, Jayaraman, S, Cunningham, K, Ciao, M, Ma, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-04 | | Release date: | 2006-05-23 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of 4-Oxalomesaconate Hydratase, LigJ, from Rhodopseudomonas palustris, Northeast Structural Genomics Target RpR66
To be Published
|
|
1ON0
 
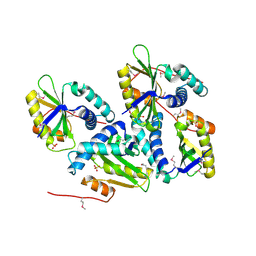 | | Crystal Structure of Putative Acetyltransferase (YycN) from Bacillus subtilis, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR144 | | Descriptor: | CHLORIDE ION, SULFATE ION, YycN protein | | Authors: | Forouhar, F, Shen, J, Kuzin, A, Chiang, Y, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-26 | | Release date: | 2003-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Putative Acetyltransferase (YycN) from Bacillus subtilis
To be Published
|
|
