2MWB
 
 | | FBP28 WW2 mutant W457F | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MWD
 
 | |
7N9T
 
 | |
7ME7
 
 | |
7MEJ
 
 | |
7MDW
 
 | |
3CZR
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) in Complex with Arylsulfonylpiperazine Inhibitor | | Descriptor: | (2R)-1-[(4-tert-butylphenyl)sulfonyl]-2-methyl-4-(4-nitrophenyl)piperazine, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Corticosteroid 11-beta-dehydrogenase isozyme 1, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2008-04-29 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and Initial SAR of Arylsulfonylpiperazine Inhibitors of 11beta-Hydroxysteroid Dehydrogenase Type 1 (11beta-HSD1)
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4OAS
 
 | | co-crystal structure of MDM2 (17-111) in complex with compound 25 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-(tert-butylsulfonyl)butan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2014-01-06 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of AMG 232, a Potent, Selective, and Orally Bioavailable MDM2-p53 Inhibitor in Clinical Development.
J.Med.Chem., 57, 2014
|
|
4KB5
 
 | | Crystal structure of MycP1 from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, Membrane-anchored mycosin mycp1 | | Authors: | Sun, D.M, He, Y, Tian, C.L. | | Deposit date: | 2013-04-23 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The putative propeptide of MycP1 in mycobacterial type VII secretion system does not inhibit protease activity but improves protein stability.
Protein Cell, 4, 2013
|
|
3PDJ
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase 1 (11b-HSD1) in Complex with 4,4-Disubstituted Cyclohexylbenzamide Inhibitor | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-[trans-4-(3-amino-3-oxopropyl)-4-phenylcyclohexyl]-N-cyclopropyl-4-[(1S)-2,2,2-trifluoro-1-hydroxy-1-methylethyl]benzamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and Optimization of Novel 4,4-Disubstituted Cyclohexylbenzamide Derivatives as Potent 11beta-HSD1 Inhibitors
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7CFS
 
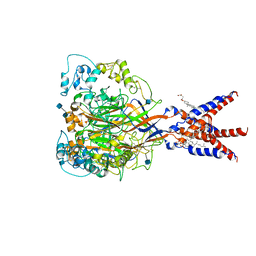 | | Cryo-EM strucutre of human acid-sensing ion channel 1a at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
7CFT
 
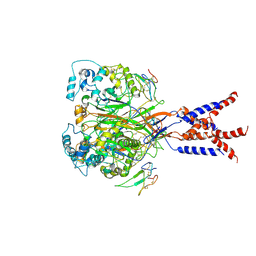 | | Cryo-EM strucutre of human acid-sensing ion channel 1a in complex with snake toxin Mambalgin1 at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Mambalgin-1 | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
3HW9
 
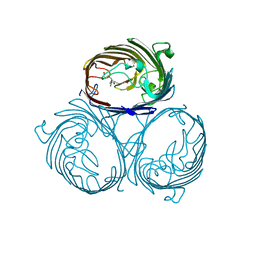 | | Cation selective pathway of OmpF porin revealed by anomalous x-ray diffraction | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Outer membrane protein F, ... | | Authors: | Balasundaresan, D, Raychaudhury, S, Blachowicz, L, Roux, B. | | Deposit date: | 2009-06-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Cation-selective pathway of OmpF porin revealed by anomalous X-ray diffraction.
J.Mol.Biol., 396, 2010
|
|
3HWB
 
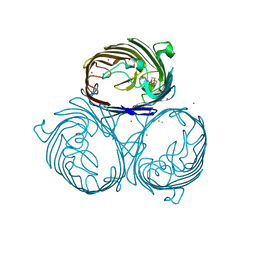 | | Cation selective pathway of OmpF porin revealed by anomalous diffraction | | Descriptor: | HEXAETHYLENE GLYCOL, Outer membrane protein F, RUBIDIUM ION | | Authors: | Balasundaresan, D, Raychaudhury, S, Blachowicz, L, Roux, B. | | Deposit date: | 2009-06-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cation-selective pathway of OmpF porin revealed by anomalous X-ray diffraction.
J.Mol.Biol., 396, 2010
|
|
4LSF
 
 | | Ion selectivity of OmpF soaked in 0.1M KBr | | Descriptor: | BROMIDE ION, GLYCEROL, Outer membrane protein F | | Authors: | Balasundaresan, D, Blachowicz, L, Roux, B. | | Deposit date: | 2013-07-22 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural study of ion permeation in OmpF porin from anomalous X-ray diffraction and molecular dynamics simulations.
J.Am.Chem.Soc., 135, 2013
|
|
4LSE
 
 | | Ion selectivity of OmpF porin soaked in 0.2M NaBr | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, BROMIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Balasundaresan, D, Blachowicz, L, Roux, B. | | Deposit date: | 2013-07-22 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural study of ion permeation in OmpF porin from anomalous X-ray diffraction and molecular dynamics simulations.
J.Am.Chem.Soc., 135, 2013
|
|
4LSH
 
 | |
4LSI
 
 | | Ion selectivity of OmpF porin soaked in 0.3M KBr | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, BROMIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Balasundaresan, D, Blachowicz, L, Roux, B. | | Deposit date: | 2013-07-22 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | A structural study of ion permeation in OmpF porin from anomalous X-ray diffraction and molecular dynamics simulations.
J.Am.Chem.Soc., 135, 2013
|
|
5DB3
 
 | | Menin in complex with MI-574 | | Descriptor: | 6-methoxy-4-methyl-1-(1H-pyrazol-4-ylmethyl)-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Borkin, D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
5DB0
 
 | | Menin in complex with MI-352 | | Descriptor: | 1-[(2R)-2,3-dihydroxypropyl]-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, 1-[(2S)-2,3-dihydroxypropyl]-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1 H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
5DB2
 
 | | Menin in complex with MI-389 | | Descriptor: | 2-{2-cyano-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indol-1-yl}aceta mide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
5DB1
 
 | | Menin in complex with MI-336 | | Descriptor: | 6-methoxy-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
5VFC
 
 | | WDR5 bound to inhibitor MM-589 | | Descriptor: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
7MBO
 
 | | FACTOR XIA (PICHIA PASTORIS; C500S [C122S]) IN COMPLEX WITH THE INHIBITOR Milvexian (BMS-986177), IUPAC NAME:(6R,10S)-10-{4-[5-chloro-2-(4-chloro-1H-1,2,3-triazol-1-yl)phenyl]-6- oxopyrimidin-1(6H)-yl}-1-(difluoromethyl)-6-methyl-1,4,7,8,9,10-hexahydro-15,11- (metheno)pyrazolo[4,3-b][1,7]diazacyclotetradecin-5(6H)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, Milvexian | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.924 Å) | | Cite: | Discovery of Milvexian, a High-Affinity, Orally Bioavailable Inhibitor of Factor XIa in Clinical Studies for Antithrombotic Therapy.
J.Med.Chem., 65, 2022
|
|
4ERE
 
 | | crystal structure of MDM2 (17-111) in complex with compound 23 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-tert-butoxy-1-oxobutan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
