1LFU
 
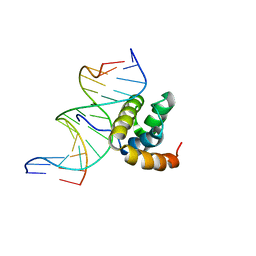 | | NMR Solution Structure of the Extended PBX Homeodomain Bound to DNA | | Descriptor: | 5'-D(*GP*CP*GP*CP*AP*TP*GP*AP*TP*TP*GP*CP*CP*C)-3', 5'-D(*GP*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*CP*GP*C)-3', homeobox protein PBX1 | | Authors: | Sprules, T, Green, N, Featherstone, M, Gehring, K. | | Deposit date: | 2002-04-12 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Lock and Key Binding of the HOX YPWM Peptide to the PBX Homeodomain
J.Biol.Chem., 278, 2003
|
|
4UFZ
 
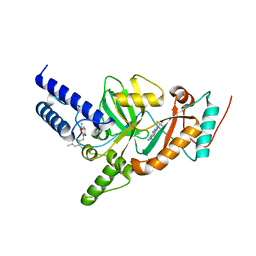 | | Synthesis of Novel NAD Dependant DNA Ligase Inhibitors via Negishi Cross-Coupling: Development of SAR and Resistance Studies | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5,7-bis(azanyl)-2-tert-butyl-4-(1,3-thiazol-2-yl)pyrido[2,3-d]pyrimidine-6-carbonitrile, DNA LIGASE | | Authors: | Murphy-Benenato, K.E, Boriack-Sjodin, P.A, Martinez-Botella, G, Carcanague, D, Gingipali, L, Gowravaram, M, Harang, J, Hale, M, Ioannidis, G, Jahic, H, Johnstone, M, Kutschke, A, Laganas, V.A, Loch, J, Oguto, H, Patel, S.J. | | Deposit date: | 2015-03-20 | | Release date: | 2015-10-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Negishi Cross-Coupling Enabled Synthesis of Novel Nad(+)-Dependent DNA Ligase Inhibitors and Sar Development.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1DU6
 
 | |
8FK6
 
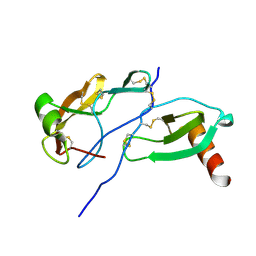 | | Crystal Structure of the Tick Evasin EVA-AAM1001(Y44A) Complexed to Human Chemokine CCL7 | | Descriptor: | C-C motif chemokine 7, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
7SCT
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL16 | | Descriptor: | C-C motif chemokine 16, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
7SCV
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL17 | | Descriptor: | C-C motif chemokine 17, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
7SCS
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL11 | | Descriptor: | Eotaxin, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
2MO7
 
 | |
2N0Q
 
 | | N2-dG-IQ modified DNA at the G1 position of the NarI recognition sequence | | Descriptor: | DNA_(5'-D(*CP*TP*CP*(IQG)P*GP*CP*GP*CP*CP*AP*TP*C)-3'), DNA_(5'-D(*GP*AP*TP*GP*GP*CP*GP*CP*CP*GP*AP*G)-3') | | Authors: | Stavros, K, Hawkins, E, Rizzo, C, Stone, M. | | Deposit date: | 2015-03-11 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Base-Displaced Intercalated Conformation of the 2-Amino-3-methylimidazo[4,5-f]quinoline N(2)-dG DNA Adduct Positioned at the Nonreiterated G(1) in the NarI Restriction Site.
Chem.Res.Toxicol., 28, 2015
|
|
2MMF
 
 | | Solution structure of AGA modified | | Descriptor: | DNA_(5'-D(*CP*TP*AP*AP*(FAG)P*AP*TP*TP*CP*A)-3'), DNA_(5'-D(*TP*GP*AP*AP*TP*CP*TP*TP*AP*G)-3') | | Authors: | Li, L, Stone, M. | | Deposit date: | 2014-03-14 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | DNA Sequence Modulates Geometrical Isomerism of the trans-8,9-Dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)-9-hydroxy Aflatoxin B1 Adduct.
Chem.Res.Toxicol., 28, 2015
|
|
2MMR
 
 | | AGC FAPY modified duplex Major isomer | | Descriptor: | DNA_(5'-D(*CP*TP*AP*AP*(FAG)P*CP*TP*TP*CP*A)-3'), DNA_(5'-D(*TP*GP*AP*AP*GP*CP*TP*TP*AP*G)-3') | | Authors: | Li, L, Stone, M. | | Deposit date: | 2014-03-17 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | DNA Sequence Modulates Geometrical Isomerism of the trans-8,9-Dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)-9-hydroxy Aflatoxin B1 Adduct.
Chem.Res.Toxicol., 28, 2015
|
|
2MMS
 
 | | AG(7-deaza)G FAPY modified duplex | | Descriptor: | DNA_(5'-D(*CP*TP*AP*AP*(FAG)P*(7GU)P*TP*TP*CP*A)-3'), DNA_(5'-D(*TP*GP*AP*AP*CP*CP*TP*TP*AP*G)-3') | | Authors: | Li, L, Stone, M. | | Deposit date: | 2014-03-17 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | DNA Sequence Modulates Geometrical Isomerism of the trans-8,9-Dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)-9-hydroxy Aflatoxin B1 Adduct.
Chem.Res.Toxicol., 28, 2015
|
|
2MMQ
 
 | | Solution structure of AGT FAPY Modified duplex | | Descriptor: | DNA_(5'-D(*CP*TP*AP*AP*(FAG)P*TP*TP*TP*CP*A)-3'), DNA_(5'-D(*TP*GP*AP*AP*AP*CP*TP*TP*AP*G)-3') | | Authors: | Li, L, Stone, M. | | Deposit date: | 2014-03-17 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | DNA Sequence Modulates Geometrical Isomerism of the trans-8,9-Dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)-9-hydroxy Aflatoxin B1 Adduct.
Chem.Res.Toxicol., 28, 2015
|
|
4NAT
 
 | | Inhibitors of 4-Phosphopanthetheine Adenylyltransferase | | Descriptor: | (1R,2R)-N-(3,4-dichlorobenzyl)-2-(4,6-dimethoxypyrimidin-2-yl)cyclohexanecarboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lahiri, S.D. | | Deposit date: | 2013-10-22 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Inhibitors of 4'-Phosphopantetheine Adenylyltransferase (PPAT) To Validate PPAT as a Target for Antibacterial Therapy.
Antimicrob.Agents Chemother., 57, 2013
|
|
4NAH
 
 | | Inhibitors of 4-Phosphopanthetheine Adenylyltransferase (PPAT) | | Descriptor: | 2-[(2-{(1S,2S)-2-[(3,4-dichlorobenzyl)carbamoyl]cyclohexyl}-6-ethylpyrimidin-4-yl)sulfanyl]-1H-imidazole-5-carboxylic acid, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Phosphopantetheine adenylyltransferase | | Authors: | Lahiri, S.D. | | Deposit date: | 2013-10-22 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of inhibitors of 4'-phosphopantetheine adenylyltransferase (PPAT) to validate PPAT as a target for antibacterial therapy.
Antimicrob.Agents Chemother., 57, 2013
|
|
4NAU
 
 | | S. aureus CoaD with Inhibitor | | Descriptor: | 2-[2-[(1S,2S)-2-[(3,4-dichlorophenyl)methylcarbamoyl]cyclohexyl]-6-ethyl-pyrimidin-4-yl]-4-oxidanyl-6-oxidanylidene-1H-pyrimidine-5-carboxamide, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Phosphopantetheine adenylyltransferase | | Authors: | Lahiri, S.D. | | Deposit date: | 2013-10-22 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of Inhibitors of 4'-Phosphopantetheine Adenylyltransferase (PPAT) To Validate PPAT as a Target for Antibacterial Therapy.
Antimicrob.Agents Chemother., 57, 2013
|
|
4J3D
 
 | | Pseudomonas aeruginosa LpxC in complex with a hydroxamate inhibitor | | Descriptor: | N~1~-hydroxy-N~5~-(3-hydroxypropyl)-N~2~-[4-(phenylethynyl)benzoyl]-L-glutamamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Lahiri, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the UDP pocket of LpxC through amino acid analogs.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
