6BVA
 
 | |
6BYH
 
 | |
6BXI
 
 | | X-ray crystal structure of NDR1 kinase domain | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase 38 | | Authors: | Xiong, S, Sicheri, F. | | Deposit date: | 2017-12-18 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Auto-Inhibition of the NDR1 Kinase Domain by an Atypically Long Activation Segment.
Structure, 26, 2018
|
|
4K25
 
 | | Crystal Structure of yeast Qri7 homodimer | | Descriptor: | CALCIUM ION, Probable tRNA threonylcarbamoyladenosine biosynthesis protein QRI7, mitochondrial, ... | | Authors: | Neculai, D, Wan, L, Mao, D.Y, Sicheri, F. | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-08 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system.
Nucleic Acids Res., 41, 2013
|
|
7KJU
 
 | | Cgi121-tRNA complex | | Descriptor: | MAGNESIUM ION, RNA (75-MER) | | Authors: | Ceccarelli, D.F, Beenstock, J, Wan, L.C.K, Sicheri, F. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | A substrate binding model for the KEOPS tRNA modifying complex.
Nat Commun, 11, 2020
|
|
7KJT
 
 | | KEOPS tRNA modifying sub-complex of archaeal Cgi121 and tRNA | | Descriptor: | RNA (70-MER), Regulatory protein Cgi121 | | Authors: | Ceccarelli, D.F, Beenstock, J, Mao, D.Y.L, Sicheri, F. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | A substrate binding model for the KEOPS tRNA modifying complex.
Nat Commun, 11, 2020
|
|
2JQZ
 
 | | Solution Structure of the C2 domain of human Smurf2 | | Descriptor: | E3 ubiquitin-protein ligase SMURF2 | | Authors: | Wiesner, S, Ogunjimi, A.A, Wang, H, Rotin, D, Sicheri, F, Wrana, J.L, Forman-Kay, J.D. | | Deposit date: | 2007-06-15 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of the HECT-Type Ubiquitin Ligase Smurf2 through Its C2 Domain
Cell(Cambridge,Mass.), 130, 2007
|
|
2KIS
 
 | | Solution structure of CA150 FF1 domain and FF1-FF2 interdomain linker | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Murphy, J.M, Hansen, D, Wiesner, S, Muhandiram, D, Borg, M, Smith, M.J, Sicheri, F, Kay, L.E, Forman-Kay, J.D, Pawson, T. | | Deposit date: | 2009-05-08 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural studies of FF domains of the transcription factor CA150 provide insights into the organization of FF domain tandem arrays.
J.Mol.Biol., 393, 2009
|
|
2K8Y
 
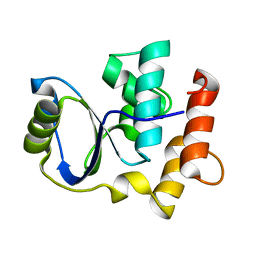 | | Solution NMR Structure of Cgi121 from Methanococcus jannaschii. Northeast Structural Genomics Consortium Target MJ0187 | | Descriptor: | Uncharacterized protein MJ0187 | | Authors: | Rumpel, S, Fares, C, Neculai, D, Arrowsmith, C, Montelione, G.T, Sicheri, F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
8T7T
 
 | |
3TWQ
 
 | |
3TWW
 
 | |
3TWS
 
 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human TERF1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, HEXAETHYLENE GLYCOL, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3TWV
 
 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human NUMA1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3LJ1
 
 | | IRE1 complexed with Cdk1/2 Inhibitor III | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Lee, K.P.K, Sicheri, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1.
Mol.Cell, 38, 2010
|
|
3LJ2
 
 | | IRE1 complexed with JAK Inhibitor I | | Descriptor: | 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Lee, K.P.K, Sicheri, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1.
Mol.Cell, 38, 2010
|
|
3LJ0
 
 | | IRE1 complexed with ADP and Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, K.P.K, Sicheri, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1.
Mol.Cell, 38, 2010
|
|
3MKS
 
 | | Crystal Structure of yeast Cdc4/Skp1 in complex with an allosteric inhibitor SCF-I2 | | Descriptor: | 1,1'-binaphthalene-2,2'-dicarboxylic acid, Cell division control protein 4, GLYCEROL, ... | | Authors: | Orlicky, S, Sicheri, F, Tyers, M, Tang, X. | | Deposit date: | 2010-04-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase.
Nat.Biotechnol., 28, 2010
|
|
1JPA
 
 | | Crystal Structure of unphosphorylated EphB2 receptor tyrosine kinase and juxtamembrane region | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, neural kinase, Nuk=Eph/Elk/Eck family receptor-like tyrosine kinase | | Authors: | Wybenga-Groot, L.E, Pawson, T, Sicheri, F. | | Deposit date: | 2001-08-01 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region.
Cell(Cambridge,Mass.), 106, 2001
|
|
2D3D
 
 | | crystal structure of the RNA binding SAM domain of saccharomyces cerevisiae Vts1 | | Descriptor: | CALCIUM ION, Vts1 protein | | Authors: | Aviv, T, Amborski, A.N, Zhao, X.S, Kwan, J.J, Johnson, P.E, Sicheri, F, Donaldson, L.W. | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The NMR and X-ray Structures of the Saccharomyces cerevisiae Vts1 SAM Domain Define a Surface for the Recognition of RNA Hairpins
J.Mol.Biol., 356, 2006
|
|
3ENP
 
 | | Crystal structure of human cgi121 | | Descriptor: | TP53RK-binding protein | | Authors: | Haffani, Y.Z, Ceccarelli, D.F, Neculai, D, Mao, D.Y, Sicheri, F. | | Deposit date: | 2008-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
1LUZ
 
 | |
1MBY
 
 | | Murine Sak Polo Domain | | Descriptor: | serine/threonine kinase | | Authors: | Leung, G.C, Hudson, J.W, Kozarova, A, Davidson, A, Dennis, J.W, Sicheri, F. | | Deposit date: | 2002-08-04 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization.
Nat.Struct.Biol., 9, 2002
|
|
1NEX
 
 | | Crystal Structure of ScSkp1-ScCdc4-CPD peptide complex | | Descriptor: | CDC4 protein, Centromere DNA-binding protein complex CBF3 subunit D, GLL(TPO)PPQSG | | Authors: | Orlicky, S, Tang, X, Willems, A, Tyers, M, Sicheri, F. | | Deposit date: | 2002-12-12 | | Release date: | 2003-02-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Phosphodependent Substrate Selection and
Orientation by the SCFCdc4 Ubiquitin Ligase
Cell(Cambridge,Mass.), 112, 2003
|
|
4I6L
 
 | |
