5Y9J
 
 | | BAFF in complex with belimumab | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, belibumab light chain, belimumab heavy chain | | Authors: | Heo, Y.-S, Shin, W. | | Deposit date: | 2017-08-25 | | Release date: | 2018-02-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | BAFF-neutralizing interaction of belimumab related to its therapeutic efficacy for treating systemic lupus erythematosus.
Nat Commun, 9, 2018
|
|
5Y9K
 
 | | Structure of the belimumab Fab fragment | | Descriptor: | belimumab heavy chain, belimumab light chain | | Authors: | Heo, Y.-S, Shin, W. | | Deposit date: | 2017-08-25 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BAFF-neutralizing interaction of belimumab related to its therapeutic efficacy for treating systemic lupus erythematosus.
Nat Commun, 9, 2018
|
|
2P0M
 
 | | Revised structure of rabbit reticulocyte 15S-lipoxygenase | | Descriptor: | (2E)-3-(2-OCT-1-YN-1-YLPHENYL)ACRYLIC ACID, Arachidonate 15-lipoxygenase, FE (II) ION | | Authors: | Choi, J, Chon, J.K, Kim, S, Shin, W. | | Deposit date: | 2007-02-28 | | Release date: | 2007-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational flexibility in mammalian 15S-lipoxygenase: Reinterpretation of the crystallographic data.
Proteins, 70, 2008
|
|
2A4V
 
 | | Crystal Structure of a truncated mutant of yeast nuclear thiol peroxidase | | Descriptor: | Peroxiredoxin DOT5 | | Authors: | Choi, J, Choi, S, Chon, J.-K, Choi, J, Cha, M.-K, Kim, I.-H, Shin, W. | | Deposit date: | 2005-06-29 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the C107S/C112S mutant of yeast nuclear 2-Cys peroxiredoxin
Proteins, 61, 2005
|
|
1QXH
 
 | |
4Y7I
 
 | | Crystal Structure of MTMR8 | | Descriptor: | Myotubularin-related protein 8, PHOSPHATE ION | | Authors: | Yoo, K, Lee, J, Son, J, Shin, W, Im, D, Heo, Y.S. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the catalytic phosphatase domain of MTMR8: implications for dimerization, membrane association and reversible oxidation.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5WUV
 
 | | Crystal structure of Certolizumab Fab | | Descriptor: | heavy chain, light chain | | Authors: | Heo, Y.S, Lee, J.U, Son, J.Y, Shin, W, Yoo, K.Y. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Int J Mol Sci, 18, 2017
|
|
1IB4
 
 | |
1IA5
 
 | | POLYGALACTURONASE FROM ASPERGILLUS ACULEATUS | | Descriptor: | POLYGALACTURONASE, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Cho, S.W, Lee, S, Shin, W. | | Deposit date: | 2001-03-22 | | Release date: | 2001-09-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of Aspergillus aculeatus polygalacturonase and a modeled structure of the polygalacturonase-octagalacturonate complex.
J.Mol.Biol., 311, 2001
|
|
1IBQ
 
 | | ASPERGILLOPEPSIN FROM ASPERGILLUS PHOENICIS | | Descriptor: | ASPERGILLOPEPSIN, ZINC ION, alpha-D-mannopyranose | | Authors: | Cho, S.W, Shin, W. | | Deposit date: | 2001-03-28 | | Release date: | 2001-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of aspergillopepsin I from Aspergillus phoenicis: variations of the S1'-S2 subsite in aspartic proteinases.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5X02
 
 | | Crystal structure of the FLT3 kinase domain bound to the inhibitor FF-10101 | | Descriptor: | N-[(2S)-1-[5-[2-[(4-cyanophenyl)amino]-4-(propylamino)pyrimidin-5-yl]pent-4-ynylamino]-1-oxidanylidene-propan-2-yl]-4-(dimethylamino)-N-methyl-but-2-enamide, Receptor-type tyrosine-protein kinase FLT3, SULFATE ION | | Authors: | Fujikawa, N, Hirano, D, Takasaki, M, Terada, D, Hagiwara, S, Park, S.-Y, Sugiyama, K. | | Deposit date: | 2017-01-19 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | A novel irreversible FLT3 inhibitor, FF-10101, shows excellent efficacy against AML cells withFLT3mutations.
Blood, 131, 2018
|
|
5X8M
 
 | | PD-L1 in complex with durvalumab | | Descriptor: | Programmed cell death 1 ligand 1, durvalumab heavy chain, durvalumab light chain | | Authors: | Heo, Y.S, Lee, H.T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
5X8L
 
 | | PD-L1 in complex with atezolizumab | | Descriptor: | Programmed cell death 1 ligand 1, atezolizumab heavy chain, atezolizumab light chain | | Authors: | Heo, Y.S, Lee, H.T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
5WUX
 
 | | TNFalpha-certolizumab Fab | | Descriptor: | Tumor necrosis factor alpha, heavy, light | | Authors: | Heo, Y.S, Lee, J.U. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Int J Mol Sci, 18, 2017
|
|
5GGS
 
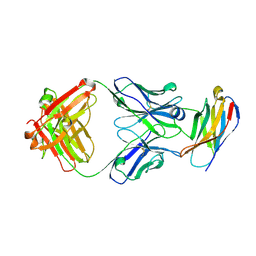 | | PD-1 in complex with pembrolizumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGV
 
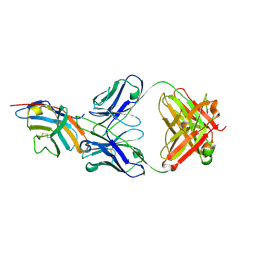 | | CTLA-4 in complex with tremelimumab Fab | | Descriptor: | Cytotoxic T-lymphocyte protein 4, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGT
 
 | | PD-L1 in complex with BMS-936559 Fab | | Descriptor: | IGK@ protein, IgG H chain, Programmed cell death 1 ligand 1 | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGR
 
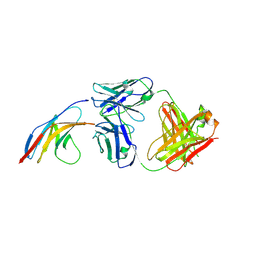 | | PD-1 in complex with nivolumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGQ
 
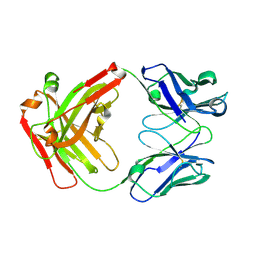 | | Crystal structure of Nivolumab Fab fragment | | Descriptor: | nivolumab heavy chain, nivolumab light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGU
 
 | | Crystal structure of tremelimumab Fab | | Descriptor: | heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
2CDV
 
 | | REFINED STRUCTURE OF CYTOCHROME C3 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Higuchi, Y, Kusunoki, M, Matsuura, Y, Yasuoka, N, Kakudo, M. | | Deposit date: | 1983-11-15 | | Release date: | 1984-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined structure of cytochrome c3 at 1.8 A resolution
J.Mol.Biol., 172, 1984
|
|
