2VV1
 
 | | hPPARgamma Ligand binding domain in complex with 4-HDHA | | Descriptor: | (4S,5E,7Z,10Z,13Z,16Z,19Z)-4-hydroxydocosa-5,7,10,13,16,19-hexaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV2
 
 | | hPPARgamma Ligand binding domain in complex with 5-HEPA | | Descriptor: | (5R,6E,8Z,11Z,14Z,17Z)-5-hydroxyicosa-6,8,11,14,17-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV3
 
 | | hPPARgamma Ligand binding domain in complex with 4-oxoDHA | | Descriptor: | (6E,10Z,13Z,16Z,19Z)-4-oxodocosa-6,10,13,16,19-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation of Ppargamma by Oxidized Fatty Acids.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV0
 
 | | hPPARgamma Ligand binding domain in complex with DHA | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2MWI
 
 | | The structure of the carboxy-terminal domain of DNTTIP1 | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1 | | Authors: | Schwabe, J.W.R, Muskett, F.W, Itoh, T. | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of a cell cycle associated HDAC1/2 complex reveals the structural basis for complex assembly and nucleosome targeting.
Nucleic Acids Res., 43, 2015
|
|
7QDT
 
 | | Crystal structure of a mutant (P393GX) Thyroid Receptor Alpha ligand binding domain designed to model dominant negative human mutations. | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Isoform Alpha-1 of Thyroid hormone receptor alpha | | Authors: | Romartinez-Alonso, B, Fairall, L, Agostini, M, Chatterjee, K, Schwabe, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Guided Approach to Relieving Transcriptional Repression in Resistance to Thyroid Hormone alpha.
Mol.Cell.Biol., 42, 2022
|
|
7ZDQ
 
 | | Cryo-EM structure of Human ACE2 bound to a high-affinity SARS CoV-2 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Bate, N, Savva, C.G, Moody, P.C.E, Brown, E.A, Schwabe, W.R, Brindle, N.P.J, Ball, J.K, Sale, J.E. | | Deposit date: | 2022-03-29 | | Release date: | 2022-05-18 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In vitro evolution predicts emerging SARS-CoV-2 mutations with high affinity for ACE2 and cross-species binding.
Plos Pathog., 18, 2022
|
|
5ICN
 
 | | HDAC1:MTA1 in complex with inositol-6-phosphate and a novel peptide inhibitor based on histone H4 | | Descriptor: | GLY-ALA-6A0-ARG-HIS, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Robertson, N.S, Watson, P.J, Jameson, A.G, Schwabe, J.W.R. | | Deposit date: | 2016-02-23 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights into the activation mechanism of class I HDAC complexes by inositol phosphates.
Nat Commun, 7, 2016
|
|
5FXY
 
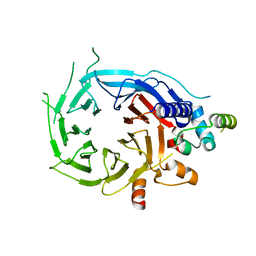 | | Structure of the human RBBP4:MTA1(464-546) complex | | Descriptor: | HISTONE-BINDING PROTEIN RBBP4, METASTASIS-ASSOCIATED PROTEIN MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
2DRP
 
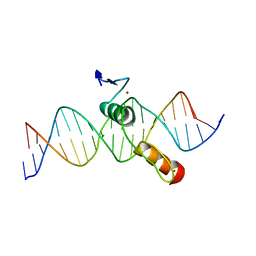 | | THE CRYSTAL STRUCTURE OF A TWO ZINC-FINGER PEPTIDE REVEALS AN EXTENSION TO THE RULES FOR ZINC-FINGER/DNA RECOGNITION | | Descriptor: | DNA (5'-D(*CP*TP*AP*AP*TP*AP*AP*GP*GP*AP*TP*AP*AP*CP*GP*TP*C P*CP*G)-3'), DNA (5'-D(*TP*CP*GP*GP*AP*CP*GP*TP*TP*AP*TP*CP*CP*TP*TP*AP*T P*TP*A)-3'), PROTEIN (TRAMTRACK DNA-BINDING DOMAIN), ... | | Authors: | Fairall, L, Schwabe, J.W.R, Chapman, L, Finch, J.T, Rhodes, D. | | Deposit date: | 1994-06-06 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition.
Nature, 366, 1993
|
|
8AU4
 
 | | Structural insights reveal a heterotetramer between oncogenic K-Ras4BG12V and Rgl2, a RalA/B activator | | Descriptor: | Ral guanine nucleotide dissociation stimulator-like 2 | | Authors: | Tariq, M, Ikeya, T, Togashi, N, Fairall, L, Alejo, C.B, Kamei, S, Alonso, B.R, Campillo, M.A.M, Hudson, A, Ito, Y, Schwabe, J, Dominguez, C, Tanaka, K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
6QPJ
 
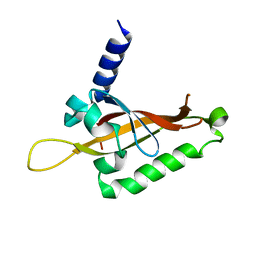 | | Human CLOCK PAS-A domain | | Descriptor: | Circadian locomoter output cycles protein kaput | | Authors: | Kwon, H, Freeman, S.L, Moody, P.C.E, Raven, E.L, Basran, J. | | Deposit date: | 2019-02-14 | | Release date: | 2019-09-25 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.315 Å) | | Cite: | Heme binding to human CLOCK affects interactions with the E-box.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1ZFD
 
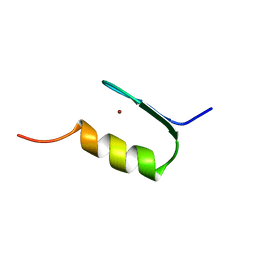 | | SWI5 ZINC FINGER DOMAIN 2, NMR, 45 STRUCTURES | | Descriptor: | SWI5, ZINC ION | | Authors: | Neuhaus, D, Nakaseko, Y, Schwabe, J.W.R, Rhodes, D, Klug, A. | | Deposit date: | 1996-04-04 | | Release date: | 1996-10-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two zinc-finger domains from SWI5 obtained using two-dimensional 1H nuclear magnetic resonance spectroscopy. A zinc-finger structure with a third strand of beta-sheet.
J.Mol.Biol., 228, 1992
|
|
2L5G
 
 | | Co-ordinates and 1H, 13C and 15N chemical shift assignments for the complex of GPS2 53-90 and SMRT 167-207 | | Descriptor: | G protein pathway suppressor 2, Putative uncharacterized protein NCOR2 | | Authors: | Oberoi, J, Yang, J, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2010-11-01 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6G16
 
 | | Structure of the human RBBP4:MTA1(464-546) complex showing loop exchange | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
7AZN
 
 | | Structure of mouse AsterC (GramD1c) with a new cholesterol-derived compound | | Descriptor: | 20alpha-hydroxy-20-(5-methylhexyl)cholesterol, ETHANOL, GLYCEROL, ... | | Authors: | Romartinez-Alonso, B, Sirvydis, K, Kim, Y, Xiao, X, Jung, M, Tontonoz, P, Schwabe, J. | | Deposit date: | 2020-11-16 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Selective Aster inhibitors distinguish vesicular and nonvesicular sterol transport mechanisms.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2YHO
 
 | | The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor | | Descriptor: | ACETATE ION, E3 UBIQUITIN-PROTEIN LIGASE MYLIP, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Fairall, L, Goult, B.T, Millard, C.J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Idol-Ube2D Complex Mediates Sterol-Dependent Degradation of the Ldl Receptor.
Genes Dev., 25, 2011
|
|
