5MAG
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
2OFU
 
 | | x-ray crystal structure of 2-aminopyrimidine carbamate 43 bound to Lck | | Descriptor: | 2,6-DIMETHYLPHENYL 2-(3,5-DIMETHOXY-4-(3-(4-METHYLPIPERAZIN-1-YL)PROPOXY)PHENYLAMINO)PYRIMIDIN- 4-YL(2,4-DIMETHOXYPHENYL)CARBAMATE, Proto-oncogene tyrosine-protein kinase LCK, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel 2-Aminopyrimidine Carbamates as Potent and Orally Active Inhibitors of Lck: Synthesis, SAR, and in Vivo Antiinflammatory Activity
J.Med.Chem., 49, 2006
|
|
4HGT
 
 | | Crystal structure of ck1d with compound 13 | | Descriptor: | 2-{2-[(3,4-difluorophenoxy)methyl]-5-methoxypyridin-4-yl}-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Casein kinase I isoform delta | | Authors: | Huang, X. | | Deposit date: | 2012-10-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Potent and Selective CK1 gamma Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
4HGL
 
 | | Crystal structure of ck1g3 with compound 1 | | Descriptor: | 2-[5-methoxy-2-(quinolin-3-yl)pyrimidin-4-yl]-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Casein kinase I isoform gamma-3, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2012-10-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Potent and Selective CK1 gamma Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
5CFB
 
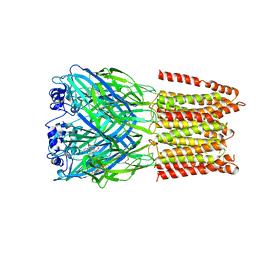 | | Crystal Structure of Human Glycine Receptor alpha-3 Bound to Strychnine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-3,Glycine receptor subunit alpha-3, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2015-07-08 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structure of human glycine receptor-alpha 3 bound to antagonist strychnine.
Nature, 526, 2015
|
|
5JM9
 
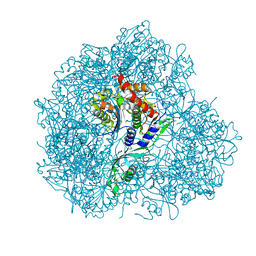 | | Structure of S. cerevesiae mApe1 dodecamer | | Descriptor: | Vacuolar aminopeptidase 1 | | Authors: | Sachse, C, Bertipaglia, C. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle.
Embo Rep., 17, 2016
|
|
3B2W
 
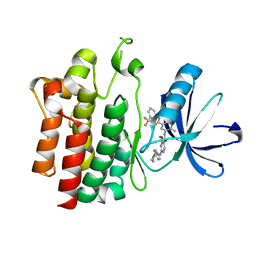 | | Crystal structure of pyrimidine amide 11 bound to Lck | | Descriptor: | N-[5-({[2-fluoro-3-(trifluoromethyl)phenyl]amino}carbonyl)-2-methylphenyl]-4-methoxy-2-[(4-piperazin-1-ylphenyl)amino]pyrimidine-5-carboxamide, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-(3-(phenylcarbamoyl)arylpyrimidine)-5-carboxamides as potent and selective inhibitors of Lck: structure, synthesis and SAR.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BYU
 
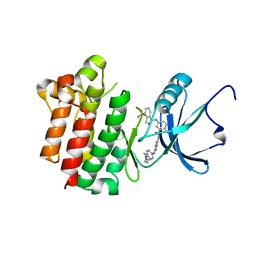 | | co-crystal structure of Lck and aminopyrimidine reverse amide 23 | | Descriptor: | 2-methyl-N-{4-methyl-3-[(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-5-yl)carbamoyl]phenyl}-3-(trifluoromethyl)benzamide, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2008-01-16 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of aminopyrimidine amides as potent, selective inhibitors of lymphocyte specific kinase: synthesis, structure-activity relationships, and inhibition of in vivo T cell activation.
J.Med.Chem., 51, 2008
|
|
3BYS
 
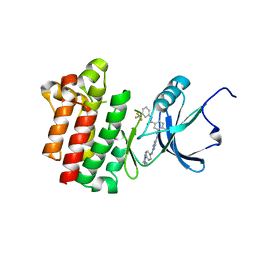 | | co-crystal structure of Lck and aminopyrimidine amide 10b | | Descriptor: | 4-methyl-N~3~-(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-5-yl)-N~1~-[3-(trifluoromethyl)phenyl]benzene-1,3-dicarboxamide, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2008-01-16 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided design of aminopyrimidine amides as potent, selective inhibitors of lymphocyte specific kinase: synthesis, structure-activity relationships, and inhibition of in vivo T cell activation.
J.Med.Chem., 51, 2008
|
|
4JV7
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JVR
 
 | | Co-crystal structure of MDM2 with inhibitor (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide | | Descriptor: | (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
3CVW
 
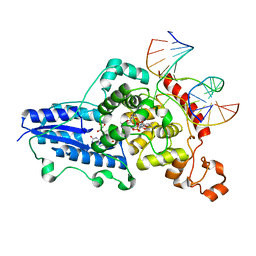 | | Drosophila melanogaster (6-4) photolyase H365N mutant bound to ds DNA with a T-T (6-4) photolesion and cofactor F0 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of a cofactor f0
accelerated (6-4) photolyase from the fruit fly
To be Published
|
|
4JVE
 
 | | Co-crystal structure of MDM2 with inhibitor (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid | | Descriptor: | (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JWR
 
 | |
3CVU
 
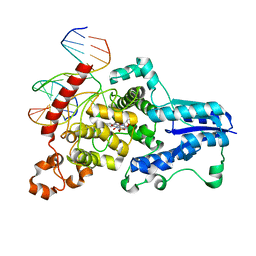 | | Drosophila melanogaster (6-4) photolyase bound to ds DNA with a T-T (6-4) photolesion | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and mechanism of a DNA (6-4) photolyase.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3CVY
 
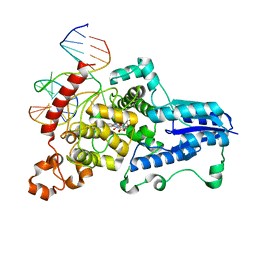 | | Drosophila melanogaster (6-4) photolyase bound to repaired ds DNA | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and mechanism of a DNA (6-4) photolyase.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
4K4F
 
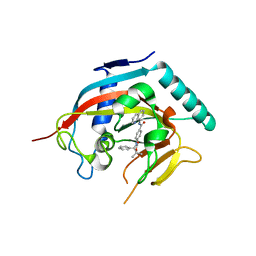 | | Co-crystal structure of TNKS1 with compound 18 [4-[(4S)-5,5-dimethyl-2-oxo-4-phenyl-1,3-oxazolidin-3-yl]-N-(quinolin-8-yl)benzamide] | | Descriptor: | 4-[(4S)-5,5-dimethyl-2-oxo-4-phenyl-1,3-oxazolidin-3-yl]-N-(quinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X, Bregman, H, Wilson, C, DiMauro, E, Gunaydin, H. | | Deposit date: | 2013-04-12 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of novel, induced-pocket binding oxazolidinones as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4K4E
 
 | | Co-crystal structure of tnks1 with compound 52 [N~2-(5-chloro-2-methoxyphenyl)-N-[trans-4-(2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)cyclohexyl]glycinamide] | | Descriptor: | N~2~-(5-chloro-2-methoxyphenyl)-N-[trans-4-(2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)cyclohexyl]glycinamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-04-12 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of novel, induced-pocket binding oxazolidinones as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4I9I
 
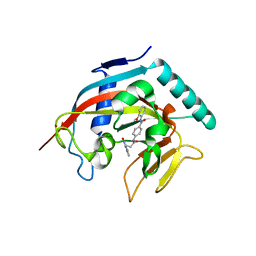 | | Crystal structure of tankyrase 1 with compound 4 | | Descriptor: | N-(2-methoxyphenyl)-4-{[3-(4-oxo-3,4-dihydroquinazolin-2-yl)propanoyl]amino}benzamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2012-12-05 | | Release date: | 2013-02-06 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a class of novel tankyrase inhibitors that bind to both the nicotinamide pocket and the induced pocket.
J.Med.Chem., 56, 2013
|
|
3CVX
 
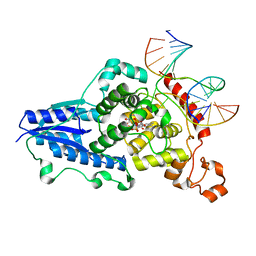 | | Drosophila melanogaster (6-4) photolyase H369M mutant bound to ds DNA with a T-T (6-4) photolesion | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of a coenzyme F0
accelerated (6-4) photolyase from the fruit fly
To be Published
|
|
3JZK
 
 | | crystal structure of MDM2 with chromenotriazolopyrimidine 1 | | Descriptor: | (6R,7S)-6,7-bis(4-bromophenyl)-7,11-dihydro-6H-chromeno[4,3-d][1,2,4]triazolo[1,5-a]pyrimidine, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and optimization of chromenotriazolopyrimidines as potent inhibitors of the mouse double minute 2-tumor protein 53 protein-protein interaction.
J.Med.Chem., 52, 2009
|
|
4JV9
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
5BK4
 
 | | Cryo-EM structure of Mcm2-7 double hexamer on dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (60-mer), strand 1, ... | | Authors: | Li, H, Yuan, Z, Bai, L. | | Deposit date: | 2017-09-12 | | Release date: | 2017-10-25 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4ERE
 
 | | crystal structure of MDM2 (17-111) in complex with compound 23 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-tert-butoxy-1-oxobutan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
4ERF
 
 | | crystal structure of MDM2 (17-111) in complex with compound 29 (AM-8553) | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S,3S)-2-hydroxypentan-3-yl]-3-methyl-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
