1G6G
 
 | |
3GMD
 
 | | Structure-based design of 7-Azaindole-pyrrolidines as inhibitors of 11beta-Hydroxysteroid-Dehydrogenase type I | | Descriptor: | 2-methyl-3-{(3S)-1-[(1-pyridin-2-ylcyclopropyl)carbonyl]pyrrolidin-3-yl}-1H-pyrrolo[2,3-b]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Valeur, E, Lepifre, F, Roche, D, Christmann-Franck, S, Hillertz, P, Musil, D. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-based design of 7-azaindole-pyrrolidine amides as inhibitors of 11 beta-hydroxysteroid dehydrogenase type I.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6F6R
 
 | | Crystal structure of human Caspase-1 with N-{3-[1-((S)-2-Hydroxy-5-oxo-tetrahydro-furan-3-ylcarbamoyl)-ethyl]-1-methyl-2,4-dioxo-1,2,3,4-tetrahydro-pyrimidin-5-yl}-4-(quinoxalin-2-ylamino)-benzamide | | Descriptor: | (3~{S})-3-[[(2~{R})-2-[3-methyl-2,6-bis(oxidanylidene)-5-[[4-(quinoxalin-2-ylamino)phenyl]carbonylamino]pyrimidin-1-yl]propanoyl]amino]-4-oxidanyl-butanoic acid, Caspase-1, SULFATE ION | | Authors: | Fournier, J.F, Clary, L, Chambon, S, Dumais, L, Harris, C.S, Millois-Barbuis, C, Pierre, R, Talano, S, Thoreau, E, Aubert, J, Aurelly, M, Bouix-Peter, C, Brethon, A, Chantalat, L, Christin, O, Comino, C, El-Bazbouz, G, Ghilini, A.L, Isabet, T, Lardy, C, Luzy, A.P, Mathieu, C, Mebrouk, K, Orfila, D, Pascau, J, Reverse, K, Roche, D, Rodeschini, V, Hennequin, L.F. | | Deposit date: | 2017-12-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Drug Design of Topically Administered Caspase 1 Inhibitors for the Treatment of Inflammatory Acne.
J. Med. Chem., 61, 2018
|
|
5MTK
 
 | | Crystal structure of human Caspase-1 with (3S,6S,10aS)-N-((2S,3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-6-(isoquinoline-1-carboxamido)-5-oxodecahydropyrrolo[1,2-a]azocine-3-carboxamide (PGE-3935199) | | Descriptor: | (3~{S})-3-[[(3~{S},6~{S},10~{a}~{S})-6-(isoquinolin-1-ylcarbonylamino)-5-oxidanylidene-2,3,6,7,8,9,10,10~{a}-octahydro-1~{H}-pyrrolo[1,2-a]azocin-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2017-01-09 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Playing against the odds: scaffold hopping from 3D-fragments
To Be Published
|
|
5MMV
 
 | | Crystal structure of human Caspase-1 with 2-((2-naphthoyl)-L-valyl)-4-hydroxy-N-((3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-2-azabicyclo[2.2.2]octane-3-carboxamide (Compound 1) | | Descriptor: | (3~{S})-3-[[(3~{S})-2-[(2~{S})-3-methyl-2-(naphthalen-2-ylcarbonylamino)butanoyl]-4-oxidanyl-2-azabicyclo[2.2.2]octan-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human Caspase-1 with 2-((2-naphthoyl)-L-valyl)-4-hydroxy-N-((3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-2-azabicyclo[2.2.2]octane-3-carboxamide (Compound 1)
To Be Published
|
|
5J26
 
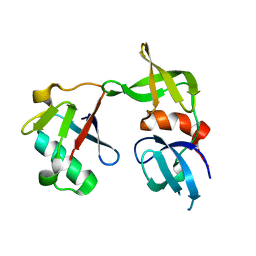 | | Crystal structure of a 53BP1 Tudor domain in complex with a ubiquitin variant | | Descriptor: | Tumor suppressor p53-binding protein 1, Ubiquitin Variant i53 | | Authors: | Wan, L, Canny, M, Juang, Y.C, Durocher, D, Sicheri, F. | | Deposit date: | 2016-03-29 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5047 Å) | | Cite: | A genetically encoded inhibitor of 53BP1 to stimulate homology-based gene editing
To Be Published
|
|
1BS5
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM | | Descriptor: | PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ZINC ION | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS8
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS4
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | Descriptor: | NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS6
 
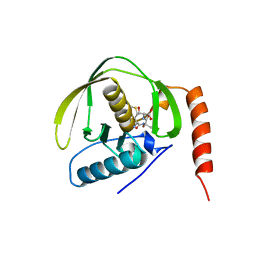 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | NICKEL (II) ION, PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BSZ
 
 | | PEPTIDE DEFORMYLASE AS FE2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | Descriptor: | FE (III) ION, NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1YJ5
 
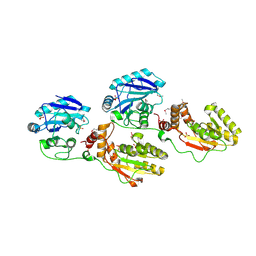 | | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme | | Descriptor: | 5' polynucleotide kinase-3' phosphatase FHA domain, 5' polynucleotide kinase-3' phosphatase catalytic domain, SULFATE ION | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-13 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
1YJM
 
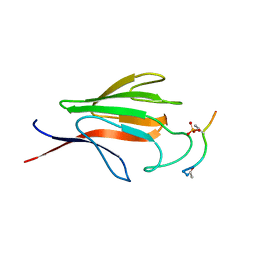 | | Crystal structure of the FHA domain of mouse polynucleotide kinase in complex with an XRCC4-derived phosphopeptide. | | Descriptor: | 12-mer peptide from DNA-repair protein XRCC4, Polynucleotide 5'-hydroxyl-kinase | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
1BS7
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM | | Descriptor: | NICKEL (II) ION, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
5KGF
 
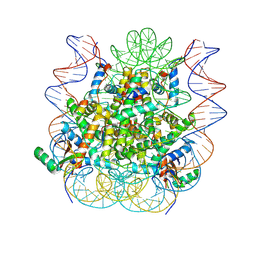 | | Structural model of 53BP1 bound to a ubiquitylated and methylated nucleosome, at 4.5 A resolution | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Wilson, M.D, Benlekbir, S, Sicheri, F, Rubinstein, J.L, Durocher, D. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | The structural basis of modified nucleosome recognition by 53BP1.
Nature, 536, 2016
|
|
1GXC
 
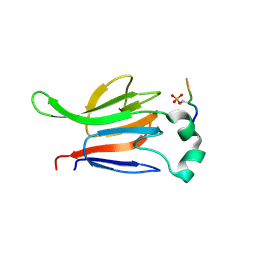 | | FHA domain from human Chk2 kinase in complex with a synthetic phosphopeptide | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE CHK2, SYNTHETIC PHOSPHOPEPTIDE | | Authors: | Li, J, Williams, B.L, Haire, L.F, Goldberg, M, Wilker, E, Durocher, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Versatility of the Fha Domain in DNA-Damage Signaling by the Tumor Suppressor Kinase Chk2
Mol.Cell, 9, 2002
|
|
6RA1
 
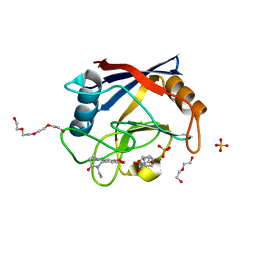 | | Human Cyclophilin D in complex with norbornane fragment derivative | | Descriptor: | 14-ethyl-4,6-dioxa-10,14-diazatricyclo[7.6.0.0^{3,7}]pentadeca-1(9),2,7-trien-13-one, 2-[(1~{R},2~{R},6~{S},7~{S})-3,5-bis(oxidanylidene)-4-azatricyclo[5.2.1.0^{2,6}]decan-4-yl]ethanoic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Graedler, U. | | Deposit date: | 2019-04-05 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6R9X
 
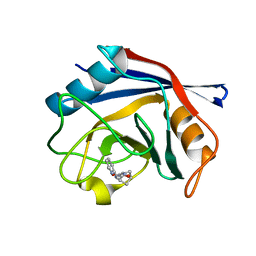 | | Human Cyclophilin D in complex with N-cyclopentyl-N'-pyridin-2-ylmethyl-oxalamide | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ~{N}'-cyclopentyl-~{N}-(pyridin-2-ylmethyl)ethanediamide | | Authors: | Graedler, U. | | Deposit date: | 2019-04-04 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8P08
 
 | | Crystal structure of human CLK1 in complex with Leucettinib-21 | | Descriptor: | (4~{Z})-4-(1,3-benzothiazol-6-ylmethylidene)-2-[[(2~{R})-1-methoxy-4-methyl-pentan-2-yl]amino]-1~{H}-imidazol-5-one, Dual specificity protein kinase CLK1 | | Authors: | Kraemer, A, Schroeder, M, Meijer, L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical, Biochemical, Cellular, and Physiological Characterization of Leucettinib-21, a Down Syndrome and Alzheimer's Disease Drug Candidate.
J.Med.Chem., 66, 2023
|
|
9FI9
 
 | | Human PIF + Z48847594 (OCCUPANCY 0.7) | | Descriptor: | ATP-dependent DNA helicase PIF1, MAGNESIUM ION, N-[4-(2-amino-1,3-thiazol-4-yl)phenyl]acetamide, ... | | Authors: | Bax, B.D, Sanders, C.M, Antson, A.A, Brandao-Neto, J. | | Deposit date: | 2024-05-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Structure-based discovery of first inhibitors targeting the helicase activity of human PIF1.
Nucleic Acids Res., 52, 2024
|
|
9FB8
 
 | | XChem refined PIF1-x0076 complex | | Descriptor: | ATP-dependent DNA helicase PIF1, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Bax, B.D, Antson, A.A, Sanders, C.M, Brandao-Neto, J. | | Deposit date: | 2024-05-12 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Structure-based discovery of first inhibitors targeting the helicase activity of human PIF1.
Nucleic Acids Res., 52, 2024
|
|
8P05
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with Leucettinib-92 | | Descriptor: | (4~{Z})-2-(1-adamantylamino)-4-(1,3-benzothiazol-6-ylmethylidene)-1~{H}-imidazol-5-one, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Meijer, L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Leucettinibs, a Class of DYRK/CLK Kinase Inhibitors Inspired by the Marine Sponge Natural Product Leucettamine B.
J.Med.Chem., 66, 2023
|
|
8P04
 
 | | Crystal structure of human CLK1 in complex with Leucettinib-92 | | Descriptor: | (4~{Z})-2-(1-adamantylamino)-4-(1,3-benzothiazol-6-ylmethylidene)-1~{H}-imidazol-5-one, Dual specificity protein kinase CLK1 | | Authors: | Kraemer, A, Schroeder, M, Meijer, L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Leucettinibs, a Class of DYRK/CLK Kinase Inhibitors Inspired by the Marine Sponge Natural Product Leucettamine B.
J.Med.Chem., 66, 2023
|
|
6R8L
 
 | | Human Cyclophilin D in complex with 1-((1S,9S,10S)-10-Hydroxy-12-oxa-8-aza-tricyclo[7.3.1.02,7]trideca-2,4,6-trien-4-ylmethyl)-3- {2-[(R)-2-(2-methylsulfanyl-phenyl)-pyrrolidin-1-yl]-2-oxo-ethyl}-urea | | Descriptor: | 1-[2-[(2~{R})-2-(2-methylsulfanylphenyl)pyrrolidin-1-yl]-2-oxidanylidene-ethyl]-3-[[(1~{S},9~{S},10~{S})-10-oxidanyl-12-oxa-8-azatricyclo[7.3.1.0^{2,7}]trideca-2(7),3,5-trien-4-yl]methyl]urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Graedler, U. | | Deposit date: | 2019-04-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6R8O
 
 | | Human Cyclophilin D in complex with 1-(((2R,3S,6R)-3-hydroxy-2,3,4,6-tetrahydro-1H-2,6-methanobenzo[c][1,5]oxazocin-8-yl)methyl)-3-(2-((R)-2-(2-(methylthio)phenyl)pyrrolidin-1-yl)-2-oxoethyl)urea | | Descriptor: | 1-(((2R,3S,6R)-3-hydroxy-2,3,4,6-tetrahydro-1H-2,6-methanobenzo[c][1,5]oxazocin-8-yl)methyl)-3-(2-((R)-2-(2-(methylthio)phenyl)pyrrolidin-1-yl)-2-oxoethyl)urea, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Graedler, U. | | Deposit date: | 2019-04-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
